| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,345,594 – 19,345,700 |
| Length | 106 |
| Max. P | 0.724374 |

| Location | 19,345,594 – 19,345,700 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.02 |
| Shannon entropy | 0.31844 |
| G+C content | 0.46744 |
| Mean single sequence MFE | -18.67 |
| Consensus MFE | -11.75 |
| Energy contribution | -11.87 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.707560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
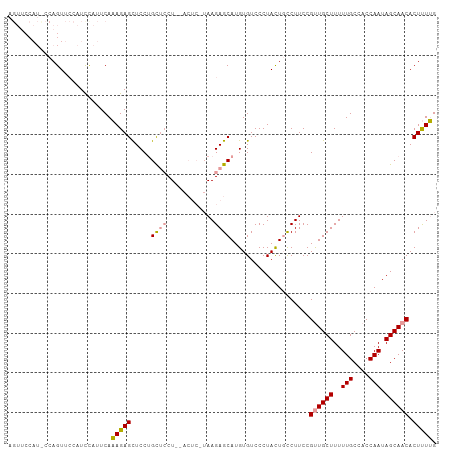
>dm3.chrX 19345594 106 + 22422827 AGUUCCAUUCCAGUUCCAUCUAUUCAAAGAGCUCCUGUUCCU--ACUCCUAAGAGCAUGUGUCCCUACUGCCUUCCGUUGCUUUUUGCCACCAAUAGCAACACUUUUG ............................((((....))))..--......(((.(((.(((....)))))))))..((((((..(((....))).))))))....... ( -15.00, z-score = -1.14, R) >droEre2.scaffold_4690 9586430 88 + 18748788 ---------CUAGUU---UCCAGUCACAGAGUUCCUGCAGCU--------AAGAGCAUGUGUCCCUAUUUUCUUCCGCUGCUUUUUGCCACCAAUAGCAACACUCUGG ---------......---........((((((....(((((.--------(((((.(((......))).)))))..)))))...((((........)))).)))))). ( -20.50, z-score = -1.66, R) >droSec1.super_8 1629606 108 + 3762037 AGUUCCAUCCCAGUUCCAUUCAUUCAAAGAGCUCCAGCUCCUCUACUCGUAAGAGCAUGUGUCCCUACUGCCUUCCGUUGCUUUUUGCCACCAAUAGCAACACUUUUG ..........((((......(((.....((((....)))).....(((....)))...))).....))))......((((((..(((....))).))))))....... ( -20.50, z-score = -2.90, R) >consensus AGUUCCAU_CCAGUUCCAUCCAUUCAAAGAGCUCCUGCUCCU__ACUC_UAAGAGCAUGUGUCCCUACUGCCUUCCGUUGCUUUUUGCCACCAAUAGCAACACUUUUG ..........................(((((.....((((............))))....................((((((..(((....))).)))))).))))). (-11.75 = -11.87 + 0.12)


| Location | 19,345,594 – 19,345,700 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.02 |
| Shannon entropy | 0.31844 |
| G+C content | 0.46744 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -18.54 |
| Energy contribution | -18.99 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.724374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
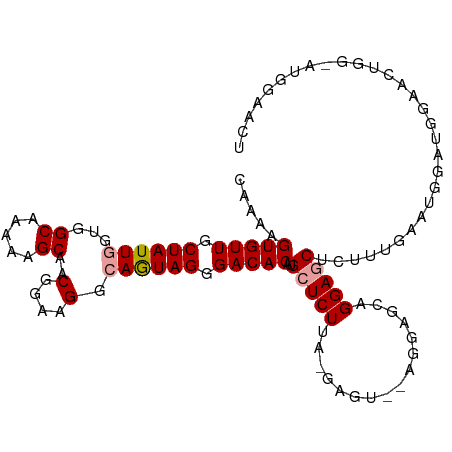
>dm3.chrX 19345594 106 - 22422827 CAAAAGUGUUGCUAUUGGUGGCAAAAAGCAACGGAAGGCAGUAGGGACACAUGCUCUUAGGAGU--AGGAACAGGAGCUCUUUGAAUAGAUGGAACUGGAAUGGAACU .....(((((.((((((...((.....))..(....).)))))).))))).(((((....))))--)....(((....(((......))).....))).......... ( -25.70, z-score = -1.50, R) >droEre2.scaffold_4690 9586430 88 - 18748788 CCAGAGUGUUGCUAUUGGUGGCAAAAAGCAGCGGAAGAAAAUAGGGACACAUGCUCUU--------AGCUGCAGGAACUCUGUGACUGGA---AACUAG--------- .((((((.((((((....))))))...(((((..((((..((.(.....)))..))))--------.)))))....)))))).....(..---..)...--------- ( -25.40, z-score = -1.37, R) >droSec1.super_8 1629606 108 - 3762037 CAAAAGUGUUGCUAUUGGUGGCAAAAAGCAACGGAAGGCAGUAGGGACACAUGCUCUUACGAGUAGAGGAGCUGGAGCUCUUUGAAUGAAUGGAACUGGGAUGGAACU .....(((((.((((((...((.....))..(....).)))))).))))).(((((....)))))(((((((....)))))))......................... ( -30.40, z-score = -2.34, R) >consensus CAAAAGUGUUGCUAUUGGUGGCAAAAAGCAACGGAAGGCAGUAGGGACACAUGCUCUUA_GAGU__AGGAGCAGGAGCUCUUUGAAUGGAUGGAACUGG_AUGGAACU .....(((((.((((((...((.....))..(....).)))))).)))))..(((((................))))).............................. (-18.54 = -18.99 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:00:54 2011