
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,342,263 – 19,342,347 |
| Length | 84 |
| Max. P | 0.610419 |

| Location | 19,342,263 – 19,342,347 |
|---|---|
| Length | 84 |
| Sequences | 10 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 75.13 |
| Shannon entropy | 0.48735 |
| G+C content | 0.45253 |
| Mean single sequence MFE | -24.63 |
| Consensus MFE | -11.18 |
| Energy contribution | -11.70 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.610419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19342263 84 - 22422827 ------GUACAUGCAUAUGCAUCCCGGUGCUGCAAAAAAGUAUGCAAAUAGAACACAAUUUGCAUGCAAAGUGCCAUCAAGAGAGCGACA---- ------((((.((((...((((....)))))))).....((((((((((........))))))))))...))))................---- ( -23.20, z-score = -1.88, R) >droYak2.chrX 9777869 83 + 21770863 ------GUACAUGCAUAUGCAUCCCAGUGCUGC-AAAAAGUAUGCAAAUAGAACACAAUUUGCAUGCAAAGUGCCAUCAAGAGAGCGACA---- ------((((.((((...((((....)))))))-)....((((((((((........))))))))))...))))................---- ( -21.70, z-score = -1.65, R) >droEre2.scaffold_4690 9583228 83 - 18748788 ------GUACAUGCAUAUGCAUCCCAGUGCUGC-AAAAAGUAUGCAAAUAGAACACAAUUUGCAUGCAAAGUGCCAUCAAGAGAGCGACA---- ------((((.((((...((((....)))))))-)....((((((((((........))))))))))...))))................---- ( -21.70, z-score = -1.65, R) >droSim1.chrX 14959671 83 - 17042790 ------GUACAUGCAUAUGCAUCCCAGUGCUGC-AAAAAGAAUGCAAAUAGAACACAAUUUGCAUGCAAAGUGCCAUCAAGAGAGCGACA---- ------((((.((((...((((....)))))))-)....(.((((((((........)))))))).)...))))................---- ( -17.10, z-score = -0.12, R) >droWil1.scaffold_181150 1675748 93 + 4952429 ACACACACACAUGUAUAAGU-GUACACUGCAUACGAAAAGUAUGCAAAUCGAGUGCAAUUUGCAUACAAAGUGCCGUCAAGAGAGACAACGAUU ......((((........))-))....(((((((.....)))))))(((((.(.(((.((((....)))).))))(((......)))..))))) ( -23.00, z-score = -2.05, R) >dp4.chrXL_group1a 3660778 86 + 9151740 ------GCACCUGGAAAGGCAGCAUAGUGCAUGCGAAAAGUAUGCAAAUUGAGUGCAAUUUGCAGGCAAAGUGCCUCCAAGGGAGCAGGCCG-- ------((.((((((..((((((((.....)))).....((.(((((((((....))))))))).))....))))))).)))..))......-- ( -30.20, z-score = -1.56, R) >droPer1.super_14 48471 86 + 2168203 ------GCACCUGGAAAUGCAGCAUAGUGCAUGCGAAAAGUAUGCAAAUUGAGUGCAAUUUGCAGGCAAAGUGCCUCCAAGGGAGCAGGCCG-- ------((.((((.(((((((.((...(((((((.....)))))))...))..))).)))).)))).....(((..(....)..))).))..-- ( -29.90, z-score = -1.50, R) >droAna3.scaffold_13417 4341529 81 + 6960332 ---------CUUGCAUAUGCAUCCCAGUGCAUGCGAAAAGUAUGCAAAUCGAGCGCAAUUUGCAUGCAAAGUGCCGUCAAGAGAGCGACA---- ---------...((((((((((....)))))).......((((((((((........))))))))))...)))).(((........))).---- ( -22.80, z-score = -0.66, R) >droVir3.scaffold_13042 4119264 86 + 5191987 ----AAACAUAUAUGCAGGGCACACAGUGCAUGCGAAAAGUAUGCAAAUCGAGUGCAAUUUGCAUGCAAAGUGGCAUCAAGAGAGCCGCG---- ----.......(((((((.((((.(..(((((((.....)))))))....).))))...)))))))....(((((.((....))))))).---- ( -30.90, z-score = -3.45, R) >droGri2.scaffold_15081 2093593 84 + 4274704 ------AUAUAUAUGCAAGACACACAGUGCAUGCGAACAGUAUGCAAAUCGAGUGCAAUUUGCAUGCAAAGUGGCACCAAGAGAGCCGCA---- ------.....((((((((.(((.(..(((((((.....)))))))....).)))...))))))))....(((((.........))))).---- ( -25.80, z-score = -2.17, R) >consensus ______GUACAUGCAUAUGCAUCACAGUGCAUGCGAAAAGUAUGCAAAUAGAGCGCAAUUUGCAUGCAAAGUGCCAUCAAGAGAGCGACA____ ............((((..((........)).........((((((((((........))))))))))...)))).................... (-11.18 = -11.70 + 0.52)
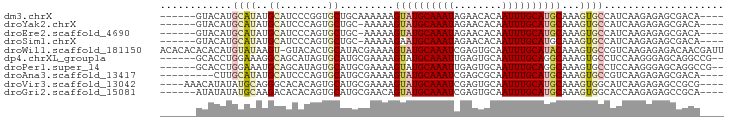


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:00:53 2011