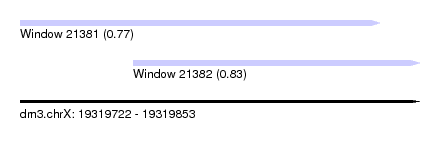
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,319,722 – 19,319,853 |
| Length | 131 |
| Max. P | 0.834907 |

| Location | 19,319,722 – 19,319,840 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 91.62 |
| Shannon entropy | 0.14180 |
| G+C content | 0.51090 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -27.78 |
| Energy contribution | -27.30 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
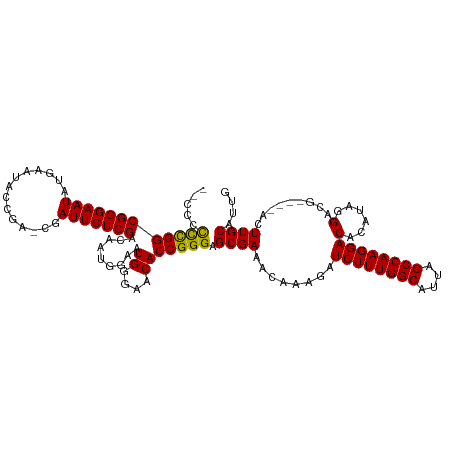
>dm3.chrX 19319722 118 + 22422827 GCCCCCUUGGCGGGAAUAUGAAUACCGACCGAUUCUCGACAAUGGAAUGGAAACACCGGGAGCGAAACAAAGAUCUUUGGCAUUAGCCAAGGACACAUAGGACAUAGCACUUGCAUUG ((.((((((((((...........))).))))...........((..((....))))))).))..........((((((((....)))))))).............((....)).... ( -31.70, z-score = -1.83, R) >droSim1.chrX 14941976 112 + 17042790 --CCCCCCGGCGGGAAUAUGAAUACCGAGCGAUUCUCGACAAUGGAAUGGGAACACCGGGAGCGAAACAAAGAUCUUUGGCAUUAGCCAAGGACACAUAGGACG----ACUUGCAUUG --...((((((((((((.((.........))))))))).........((....))))))).((((........((((((((....))))))))......(....----.))))).... ( -29.20, z-score = -0.89, R) >droSec1.super_8 1611728 112 + 3762037 --CCCCCCGGCGGGAAUAUGAAUACCGAGCGAUUCUCGACAAUGGAAUGGGAACACCGGGAGCGAAACAAAGAUCUUUGGCAUUAGCCAAGGACAUACAGGACG----ACUUGCAUUG --...((((((((((((.((.........))))))))).........((....))))))).((((........((((((((....))))))))......(....----.))))).... ( -29.20, z-score = -0.89, R) >droYak2.chrX 9761986 110 - 21770863 GCCCCCCCGGCGGGAAUAUGAAUACC----GAUUCUCGACAAUGGAAUGGGAACACCGGGAGCGAAACAAAGAUCUUUGGCAUUAGCCAAGGACACACAGGACG----ACUUGCAUUG ((.((((((.(((((((.((.....)----))))))))....)))...((.....))))).))..........((((((((....))))))))....((((...----.))))..... ( -31.40, z-score = -1.59, R) >droEre2.scaffold_4690 9568873 108 + 18748788 --CCCUCCGGCGGGAAUAUGAAUACG----GAUUCUCGACAAUGGAAUGGGAACACCGGGAGCGAAACAAAGAUCUUUGGCAUUAGCCAAGGACACAUCAGACG----ACUUGCAUUG --(((((((.(((((((.((....))----.)))))))....))))..((.....))))).((((........((((((((....))))))))....((....)----).)))).... ( -31.10, z-score = -2.18, R) >consensus __CCCCCCGGCGGGAAUAUGAAUACCGA_CGAUUCUCGACAAUGGAAUGGGAACACCGGGAGCGAAACAAAGAUCUUUGGCAUUAGCCAAGGACACAUAGGACG____ACUUGCAUUG .....((((((((((((..............))))))).........((....))))))).((((........((((((((....))))))))(......).........)))).... (-27.78 = -27.30 + -0.48)


| Location | 19,319,759 – 19,319,853 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.32 |
| Shannon entropy | 0.30565 |
| G+C content | 0.50462 |
| Mean single sequence MFE | -26.39 |
| Consensus MFE | -19.34 |
| Energy contribution | -19.34 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
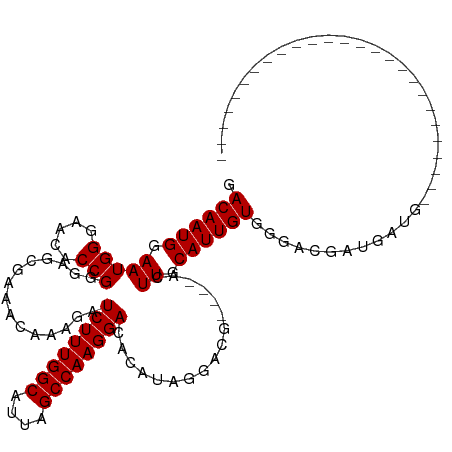
>dm3.chrX 19319759 94 + 22422827 GACAAUGGAAUGGAAACACCGGGAGCGAAACAAAGAUCUUUGGCAUUAGCCAAGGACACAUAGGACAUAGCACUUGCAUUGUGGCAGUGCGAUG-------------------------- (....(((..((....)))))....)..........((((((((....)))))))).........(((.(((((.((......))))))).)))-------------------------- ( -25.30, z-score = -2.22, R) >droSim1.chrX 14942011 90 + 17042790 GACAAUGGAAUGGGAACACCGGGAGCGAAACAAAGAUCUUUGGCAUUAGCCAAGGACACAUAGGACG----ACUUGCAUUGUGGGACGAUGAUG-------------------------- .((((((...(((.....))).(((((.........((((((((....)))))))).........))----.))).))))))............-------------------------- ( -20.87, z-score = -1.92, R) >droSec1.super_8 1611763 90 + 3762037 GACAAUGGAAUGGGAACACCGGGAGCGAAACAAAGAUCUUUGGCAUUAGCCAAGGACAUACAGGACG----ACUUGCAUUGUGGGACGAUGAUG-------------------------- .((((((...(((.....))).(((((.........((((((((....)))))))).........))----.))).))))))............-------------------------- ( -20.87, z-score = -1.92, R) >droYak2.chrX 9762019 116 - 21770863 GACAAUGGAAUGGGAACACCGGGAGCGAAACAAAGAUCUUUGGCAUUAGCCAAGGACACACAGGACG----ACUUGCAUUGUGGAACGAUGAUGGCGACCUCCUGGCUGGCACUGCGAUG ...........((...(((((((((.(.........((((((((....)))))))).........((----.((..(((((.....)))))..)))).)))))))).))...))...... ( -31.50, z-score = -1.11, R) >droEre2.scaffold_4690 9568904 109 + 18748788 GACAAUGGAAUGGGAACACCGGGAGCGAAACAAAGAUCUUUGGCAUUAGCCAAGGACACAUCAGACG----ACUUGCAUUGUGGGACGGUGAUGGCGACCA-------GGCAGUGCGUUG ..(((((.(.((((....))((..((..........((((((((....))))))))..(((((..((----.((..(...)..)).)).)))))))..)).-------..)).).))))) ( -33.40, z-score = -1.82, R) >consensus GACAAUGGAAUGGGAACACCGGGAGCGAAACAAAGAUCUUUGGCAUUAGCCAAGGACACAUAGGACG____ACUUGCAUUGUGGGACGAUGAUG__________________________ .((((((.(((((.....)))...............((((((((....)))))))).................)).))))))...................................... (-19.34 = -19.34 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:00:46 2011