| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,312,966 – 19,313,187 |
| Length | 221 |
| Max. P | 0.948161 |

| Location | 19,312,966 – 19,313,084 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.69 |
| Shannon entropy | 0.33662 |
| G+C content | 0.57272 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.43 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.713810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19312966 118 - 22422827 --UUUUGUUUUGGCCAUCGCCGAACUGAUGGCGGAUGGUGAUGUCACCGGUGAUUCCUCGAUGCUUCUGACAACGCCUCCCCCCUUCAUAGAACCACCACCCGCACCCCCCCCCCCCCCC --....(((((((((((((......)))))))(((.((((.(((((..(((...........)))..))))).)))))))........)))))).......................... ( -29.20, z-score = -1.68, R) >droSim1.chrX 14935122 99 - 17042790 --UUUUGUUUUGGCCAUCGCCGAACUGAUGGCGGAUGGUGAUGUCACCGGUGAUUCCUCGAUGCUUCUGACAAC-CAUCCCCCCCUCACAUACCCUUCACCC------------------ --..........(((((((......)))))))(((((((..(((((..(((...........)))..)))))))-)))))......................------------------ ( -26.90, z-score = -1.54, R) >droSec1.super_8 1604743 98 - 3762037 --UUUUGUUUUGGCCAUCGCCGAACUGAUGGCGGAUGGUGAUGUCACCGGUGAUUCCUCGAUGCUUCUGACAAC-CAUCCCCCCUCCCCAC-CCCUUCACCC------------------ --..........(((((((......)))))))(((((((..(((((..(((...........)))..)))))))-)))))...........-..........------------------ ( -26.90, z-score = -1.67, R) >droYak2.chrX 9754715 94 + 21770863 AUUUUUGUUUUGGCCAUCGCCGUACCGAUGGCGGAUGGUGAUGUCACCGGUGAUUCCUCGAUGCUUCUGACAACAAACCCCCCUCCCCCCGCGA-------------------------- ....(((((..(((.((((((((....)))))((((((((....))))).....)))..))))))...))))).....................-------------------------- ( -23.30, z-score = 0.14, R) >consensus __UUUUGUUUUGGCCAUCGCCGAACUGAUGGCGGAUGGUGAUGUCACCGGUGAUUCCUCGAUGCUUCUGACAAC_CAUCCCCCCCCCACAGACCCUUCACCC__________________ ....(((((..(((.(((((((......))))((((((((....))))).....)))..))))))...)))))............................................... (-21.42 = -21.43 + 0.00)


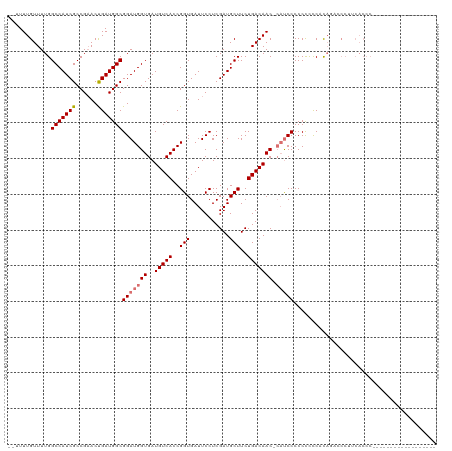
| Location | 19,313,006 – 19,313,108 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.92 |
| Shannon entropy | 0.21939 |
| G+C content | 0.50970 |
| Mean single sequence MFE | -31.96 |
| Consensus MFE | -27.76 |
| Energy contribution | -27.96 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19313006 102 + 22422827 GGAGGCGUUGUCAGAAGCAUCGAGGAAUCACCGGUGACAUCACCAUCCGCCAUCAGUUCGGCGAUGGCCAAAACAAAAAUAAACG------------------UCGGAGUCUGGCAAUCG ......(((((((((....((((.(.......((((....))))....((((((........))))))...............).------------------))))..))))))))).. ( -31.30, z-score = -1.58, R) >droSim1.chrX 14935144 101 + 17042790 -GGAUGGUUGUCAGAAGCAUCGAGGAAUCACCGGUGACAUCACCAUCCGCCAUCAGUUCGGCGAUGGCCAAAACAAAAAUAAACG------------------CUGGAGUCUGGCAAUCG -....((((((((((....((.((........((((....))))....((((((........)))))).................------------------)).)).)))))))))). ( -33.50, z-score = -2.29, R) >droSec1.super_8 1604764 101 + 3762037 -GGAUGGUUGUCAGAAGCAUCGAGGAAUCACCGGUGACAUCACCAUCCGCCAUCAGUUCGGCGAUGGCCAAAACAAAAAUAAACG------------------CUGGAGUCUGGCAAUCG -....((((((((((....((.((........((((....))))....((((((........)))))).................------------------)).)).)))))))))). ( -33.50, z-score = -2.29, R) >droYak2.chrX 9754730 119 - 21770863 -GGUUUGUUGUCAGAAGCAUCGAGGAAUCACCGGUGACAUCACCAUCCGCCAUCGGUACGGCGAUGGCCAAAACAAAAAUAAACGAAUAAACGAAUAUAAACGCCGGAGUCUGGCAAUCG -.....(((((((((......((....)).((((((............(((((((......)))))))...............((......))........))))))..))))))))).. ( -33.60, z-score = -1.68, R) >droEre2.scaffold_4690 9562342 96 + 18748788 ------CUUGUCAGAAGCAUCAAGGAAUCACCGGUGACAUCACCAUCCGCCAUCAGUUCGGCGAUGGCCAAAACAAAAUUAAACG------------------CCGGAGUCUGGCAAUCG ------.((((((((..(.....((.......((((....))))....((((((........)))))).................------------------)).)..))))))))... ( -27.90, z-score = -2.13, R) >consensus _GGAUGGUUGUCAGAAGCAUCGAGGAAUCACCGGUGACAUCACCAUCCGCCAUCAGUUCGGCGAUGGCCAAAACAAAAAUAAACG__________________CCGGAGUCUGGCAAUCG ......(((((((((..(.....((.....))((((....))))....((((((........))))))......................................)..))))))))).. (-27.76 = -27.96 + 0.20)

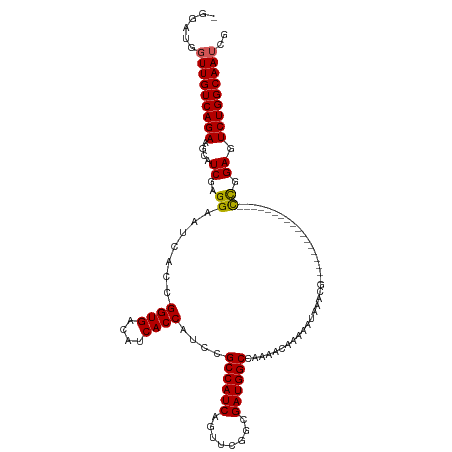
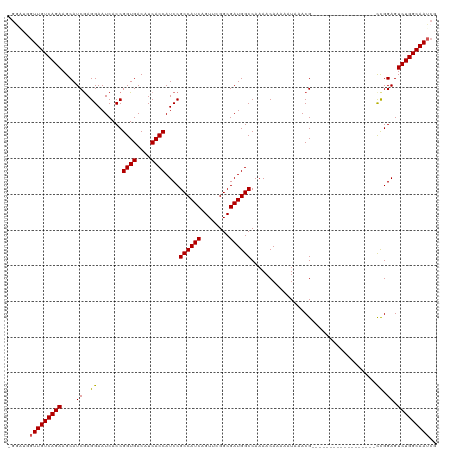
| Location | 19,313,084 – 19,313,187 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.48 |
| Shannon entropy | 0.19960 |
| G+C content | 0.50625 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -29.48 |
| Energy contribution | -30.16 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chrX 19313084 103 - 22422827 -AGGAAGGAGUGAGGGAGAUGGUGCCGCCAGAUUCUGAUUUCGUUCGUCAUUUUGAACUUUGUCGGUUCAGCGCGCCCAUCGAUUGCCAGACUCCGACGUUUAU---------------- -.....(((((...((.(((((.(((((..((..(((((...(((((......)))))...))))).)).))).))))))).....))..))))).........---------------- ( -34.80, z-score = -1.35, R) >droSim1.chrX 14935221 102 - 17042790 -AGGAAGGAGUGAGGGAGAUGGUGCCGCCAGAUUCUGAUUUCGUUCGUCA-UUUGAACUUUGUCGGUUCAGCGCGCCCAUCGAUUGCCAGACUCCAGCGUUUAU---------------- -.....(((((...((.(((((.(((((..((..(((((...(((((...-..)))))...))))).)).))).))))))).....))..))))).........---------------- ( -34.60, z-score = -1.40, R) >droSec1.super_8 1604841 102 - 3762037 -AGGAAGGAGUGAGGAAGAUGGUGCCGCCAGAUUCUGAUUUCGUUCGUCA-UUUGAACUUUGUCGGUUCAGCGCGCCCAUCGAUUGCCAGACUCCAGCGUUUAU---------------- -.....(((((..((..(((((.(((((..((..(((((...(((((...-..)))))...))))).)).))).))))))).....))..))))).........---------------- ( -35.40, z-score = -1.96, R) >droYak2.chrX 9754809 112 + 21770863 -AGGAAGGAGU------GAUGGUGCCGCCAGAUUCUGAUUUCGUUCGUCA-UUUGAACUUUGUCGGUUCAGCGCGCCCAUCGAUUGCCAGACUCCGGCGUUUAUAUUCGUUUAUUCGUUU -..(((.((((------(((((.(((((..((..(((((...(((((...-..)))))...))))).)).))).)))))))....(((.......)))......)))).)))........ ( -32.80, z-score = -0.97, R) >droEre2.scaffold_4690 9562414 97 - 18748788 AAGGAAGGAGU------GAUGGUGUCGCCAGAUUCUGAUUUCGUUCGUCA-UUUGAACUUUGCUGGUUCAGCGCGCCCAUCGAUUGCCAGACUCCGGCGUUUAA---------------- ...(..(((((------(((((.(.(((..............(((((...-..)))))...((((...)))))))))))))(.....)..)))))..)......---------------- ( -25.60, z-score = 0.64, R) >consensus _AGGAAGGAGUGAGG_AGAUGGUGCCGCCAGAUUCUGAUUUCGUUCGUCA_UUUGAACUUUGUCGGUUCAGCGCGCCCAUCGAUUGCCAGACUCCGGCGUUUAU________________ ......(((((..((..(((((.(((((..((..(((((...(((((......)))))...))))).)).))).))))))).....))..)))))......................... (-29.48 = -30.16 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:00:42 2011