| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,005,411 – 12,005,491 |
| Length | 80 |
| Max. P | 0.787724 |

| Location | 12,005,411 – 12,005,491 |
|---|---|
| Length | 80 |
| Sequences | 7 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 68.82 |
| Shannon entropy | 0.56716 |
| G+C content | 0.51743 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -9.84 |
| Energy contribution | -11.36 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.787724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12005411 80 - 23011544 GUGCCAGGGUCAGUUGAACGUUAACUGGCCCUUUCGUUUGGAGUGUAUUUGGUUCAAGGCU---GUUCUCAUGGC-CCCCAGCC ..(((((((((((((((...)))))))))))).....(((((.(......).)))))))).---........(((-.....))) ( -25.30, z-score = -0.92, R) >droPer1.super_8 2941809 75 - 3966273 ------CGUUCAGGGUUACGUAUUUCG---UGUUCAUUUAUUUUAUGUGCAGUUCAAGGCUAUCAUCCGCCCCGUCCCCCUGUU ------....(((((..(((.......---(((.(((.......))).)))......(((........))).)))..))))).. ( -17.40, z-score = -3.09, R) >dp4.chr4_group2 280889 75 + 1235136 ------CGUUCAGGGUUACGUAUUUUG---UGUUCCUUUAUUUUAUGUGCAGUUCAAGGCUAUCAUCCGCCCCGUCCCCCCGUU ------......(((..(((....(((---..(.............)..))).....(((........))).)))..))).... ( -11.92, z-score = -0.66, R) >droEre2.scaffold_4929 13232272 80 + 26641161 GUGCCAGGGUCACUUUGACCUUAACUGGCCCUUUCGCUUGGAGCGUAUUUGGUUCAAGGCU---GUUCUCAUGGC-CCGCAGCC ..(((((((((.....))))....))))).......((((((.((....)).))))))(((---((((....)).-..))))). ( -24.10, z-score = 0.06, R) >droYak2.chr2L 8424374 79 - 22324452 GUGCCAGGGCCAGUUUGACGUUAACUGGCCCUUUCGUUUGGAGCGUAUAUAGUUCAAGGCU---GUUCUCAUGGC-CCCCAGC- ..(((((((((((((.......)))))))))....((((.((((.......)))).)))).---.......))))-.......- ( -28.30, z-score = -2.04, R) >droSec1.super_16 200958 81 - 1878335 GUUCCAGGGUCAGUUGAACGUUAACUGGCCCUUUCAUUUGGAGCGUAUUUGGUUCAAGGCU---GUUCUCAUGGUCCCCCAGCC (((((((((((((((((...))))))))))).......)))))).............((((---(..............))))) ( -26.75, z-score = -2.04, R) >droSim1.chr2L 11809545 80 - 22036055 GUUCCAGGGUCAGUUGAACGUUAACUGGCCCUUUCGUUUGGAGCGUAUUUGGUUCAAGGCU---GUUCUCAUGGU-CCACAGCC (((((((((((((((((...))))))))))).......)))))).............((((---((((....)).-..)))))) ( -28.51, z-score = -2.83, R) >consensus GUGCCAGGGUCAGUUUAACGUUAACUGGCCCUUUCGUUUGGAGCGUAUUUGGUUCAAGGCU___GUUCUCAUGGU_CCCCAGCC .....(((((((((((.....)))))))))))....((((((.((....)).)))))).......................... ( -9.84 = -11.36 + 1.52)

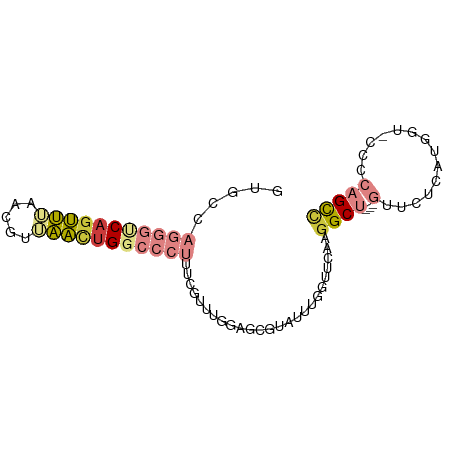
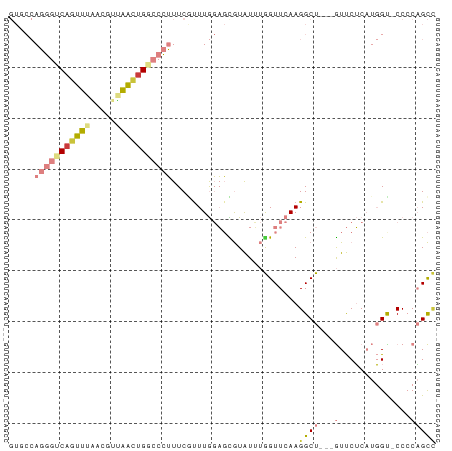
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:09 2011