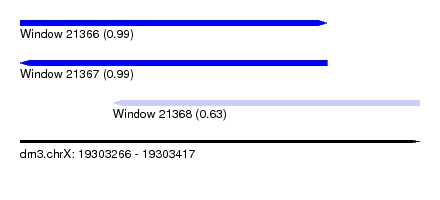
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,303,266 – 19,303,417 |
| Length | 151 |
| Max. P | 0.994682 |

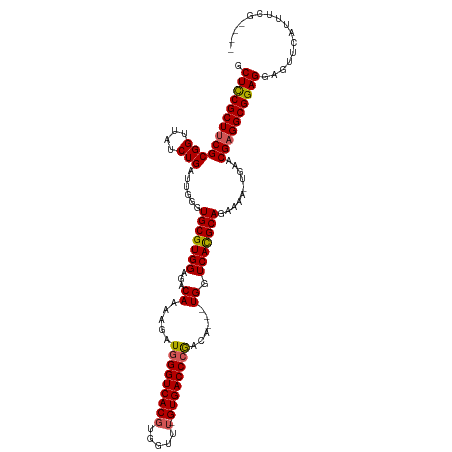
| Location | 19,303,266 – 19,303,382 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.17 |
| Shannon entropy | 0.16805 |
| G+C content | 0.53452 |
| Mean single sequence MFE | -43.23 |
| Consensus MFE | -40.88 |
| Energy contribution | -40.80 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.994682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
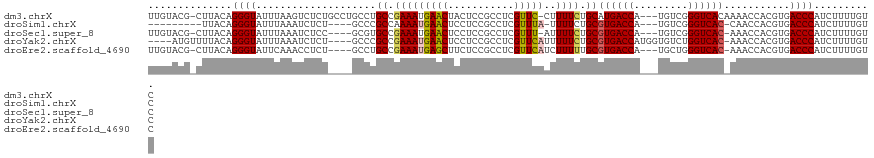
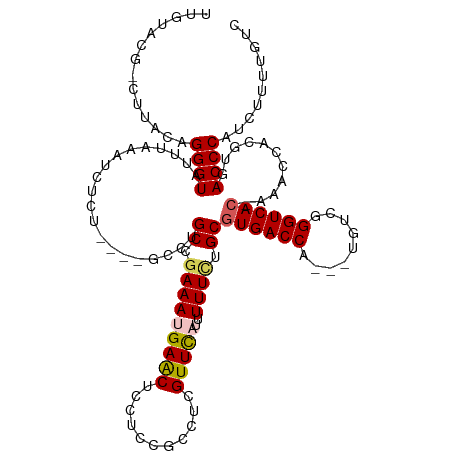
>dm3.chrX 19303266 116 + 22422827 GCUCCGCUUCGCGGUUAUCUGAUUGGGUGCGUGGAGACAAAAGAUGGGUCACGUGGUUUUGUGACCCGACA---UGGUCAUGCAGAAAA-GGAACGAGGCGGAGUAGUUCAUUUCGGCAG ((((((((((((((((....)))))..(((((((...((...(.(((((((((......))))))))).).---)).))))))).....-....)))))))))))............... ( -43.90, z-score = -2.59, R) >droSim1.chrX 14920786 111 + 17042790 GCUCCGCUUCGCGGUUAUCUGAUUGGGUGCGUGGAGACAAAAGAUGGGUCACGUGGUUG-GUGACCCGACA---UGGUCACGCAGAAAA-UAAACGAGGCGGAGGAGUUCAUUUUG---- .(((((((((((((((....)))))..(((((((...((...(.((((((((.......-)))))))).).---)).))))))).....-....))))))))))............---- ( -43.10, z-score = -3.06, R) >droSec1.super_8 1595268 111 + 3762037 GCUCCGCUUCGCGGUUAUCUGAUUGGGUGCGUGGAGACAAAAGAUGGGUCACGUGGUUU-GUGACCCGACA---UGGUCACGCAGAAAA-UAAACGAGGCGGAGGAGUUCAUUUCG---- .(((((((((((((((....)))))..(((((((...((...(.(((((((((.....)-)))))))).).---)).))))))).....-....))))))))))(((.....))).---- ( -44.50, z-score = -3.32, R) >droYak2.chrX 9744995 115 - 21770863 GCUCCGCUACGCGGUUAUCUGGUUGGAUGCGUGGAGACAAAAGAUGGGUCACGUGGUUU-GUGACCAGACACCAUGGUCACGCAGAAAAAUGAACGAGGCGGAGGAGUUCAUUUCG---- .(((((((.(((((....)))(((...(((((((...((.......(((((((.....)-))))))........)).)))))))....)))...)).)))))))(((.....))).---- ( -37.56, z-score = -1.02, R) >droEre2.scaffold_4690 9553284 112 + 18748788 GCUUCGCUUCGCGGUUGUCUGGUUGGGUGCGUGGAGACAAAAGAUGGGUCACGUGGUUU-GUGACCCAGCA---UGGUCACGCAAAAAGAUGAACGAGGCGGAGAAGCUCAUUUCG---- .((((((((((...(..(((..(((.((((.((....))...(.(((((((((.....)-)))))))).).---..).))).)))..)))..).))))))))))............---- ( -47.10, z-score = -3.48, R) >consensus GCUCCGCUUCGCGGUUAUCUGAUUGGGUGCGUGGAGACAAAAGAUGGGUCACGUGGUUU_GUGACCCGACA___UGGUCACGCAGAAAA_UGAACGAGGCGGAGGAGUUCAUUUCG____ .(((((((((((((....)))......(((((((...((.....((((((((........))))))))......)).)))))))..........))))))))))................ (-40.88 = -40.80 + -0.08)

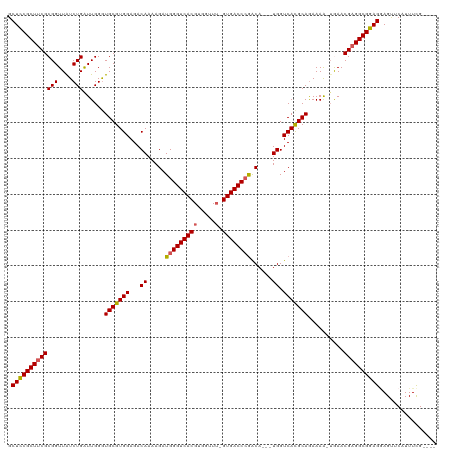
| Location | 19,303,266 – 19,303,382 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.17 |
| Shannon entropy | 0.16805 |
| G+C content | 0.53452 |
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -28.19 |
| Energy contribution | -28.99 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.992372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19303266 116 - 22422827 CUGCCGAAAUGAACUACUCCGCCUCGUUCC-UUUUCUGCAUGACCA---UGUCGGGUCACAAAACCACGUGACCCAUCUUUUGUCUCCACGCACCCAAUCAGAUAACCGCGAAGCGGAGC ................((((((.((((...-...((((...(((..---.)))(((((((........)))))))........................)))).....)))).)))))). ( -33.40, z-score = -3.11, R) >droSim1.chrX 14920786 111 - 17042790 ----CAAAAUGAACUCCUCCGCCUCGUUUA-UUUUCUGCGUGACCA---UGUCGGGUCAC-CAACCACGUGACCCAUCUUUUGUCUCCACGCACCCAAUCAGAUAACCGCGAAGCGGAGC ----............((((((.(((((((-(((..((((((..((---.(..(((((((-.......)))))))..)...))....)))))).......))))))..)))).)))))). ( -35.40, z-score = -4.10, R) >droSec1.super_8 1595268 111 - 3762037 ----CGAAAUGAACUCCUCCGCCUCGUUUA-UUUUCUGCGUGACCA---UGUCGGGUCAC-AAACCACGUGACCCAUCUUUUGUCUCCACGCACCCAAUCAGAUAACCGCGAAGCGGAGC ----............((((((.(((((((-(((..((((((..((---.(..(((((((-.......)))))))..)...))....)))))).......))))))..)))).)))))). ( -36.00, z-score = -3.81, R) >droYak2.chrX 9744995 115 + 21770863 ----CGAAAUGAACUCCUCCGCCUCGUUCAUUUUUCUGCGUGACCAUGGUGUCUGGUCAC-AAACCACGUGACCCAUCUUUUGUCUCCACGCAUCCAACCAGAUAACCGCGUAGCGGAGC ----............((((((..(((..........(((((..((.((((...((((((-.......))))))))))...))....)))))(((......)))....)))..)))))). ( -32.80, z-score = -2.23, R) >droEre2.scaffold_4690 9553284 112 - 18748788 ----CGAAAUGAGCUUCUCCGCCUCGUUCAUCUUUUUGCGUGACCA---UGCUGGGUCAC-AAACCACGUGACCCAUCUUUUGUCUCCACGCACCCAACCAGACAACCGCGAAGCGAAGC ----........(((((...((.((((...(((...((((((..((---.(.((((((((-.......)))))))).)...))....)))))).......))).....)))).))))))) ( -30.70, z-score = -2.05, R) >consensus ____CGAAAUGAACUCCUCCGCCUCGUUCA_UUUUCUGCGUGACCA___UGUCGGGUCAC_AAACCACGUGACCCAUCUUUUGUCUCCACGCACCCAAUCAGAUAACCGCGAAGCGGAGC ................((((((.((((.........((((((........(..(((((((........)))))))..).........))))))........(.....))))).)))))). (-28.19 = -28.99 + 0.80)

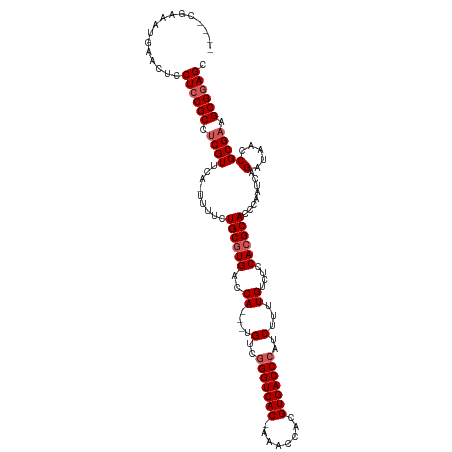
| Location | 19,303,301 – 19,303,417 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 85.06 |
| Shannon entropy | 0.25520 |
| G+C content | 0.48730 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -18.72 |
| Energy contribution | -18.60 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.631536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19303301 116 - 22422827 UUGUACG-CUUACAGGGUAUUUAAGUCUCUGCCUGCCUGCCGAAAUGAACUACUCCGCCUCGUUC-CUUUUCUGCAUGACCA---UGUCGGGUCACAAAACCACGUGACCCAUCUUUUGUC ......(-(...(((((.........)))))...)).(((.((((.((((...........))))-..)))).))).(((..---.)))(((((((........))))))).......... ( -27.80, z-score = -1.49, R) >droSim1.chrX 14920821 103 - 17042790 ---------UUACAGGGUAUUUAAAUCUCU----GCCCGCCAAAAUGAACUCCUCCGCCUCGUUUA-UUUUCUGCGUGACCA---UGUCGGGUCAC-CAACCACGUGACCCAUCUUUUGUC ---------((((.(((((..........)----))))((.(((((((((...........)))))-))))..))))))...---....(((((((-.......))))))).......... ( -26.60, z-score = -2.85, R) >droSec1.super_8 1595303 111 - 3762037 UUGUACG-CUUACAGGGUAUUUAAAUCUCC----GCGUGCCGAAAUGAACUCCUCCGCCUCGUUU-AUUUUCUGCGUGACCA---UGUCGGGUCAC-AAACCACGUGACCCAUCUUUUGUC .((((..-..))))((((...........(----(((((..(((((((((...........))))-)))))....((((((.---.....))))))-....)))))))))).......... ( -27.10, z-score = -1.29, R) >droYak2.chrX 9745030 112 + 21770863 ----AUGUUUUACAGGGUAUUUAAAUCUCU----GCCCGCCGAAAUGAACUCCUCCGCCUCGUUCAUUUUUCUGCGUGACCAUGGUGUCUGGUCAC-AAACCACGUGACCCAUCUUUUGUC ----(((..((((.(((((..........)----))))((.(((((((((...........)))))))))...))(((((((.......)))))))-.......))))..)))........ ( -28.80, z-score = -2.38, R) >droEre2.scaffold_4690 9553319 112 - 18748788 UUGUACG-CUUACAGGGUAUUCAAACCUCU----GCCUGCCGAAAUGAGCUUCUCCGCCUCGUUCAUCUUUUUGCGUGACCA---UGCUGGGUCAC-AAACCACGUGACCCAUCUUUUGUC ....(((-(...((((((......))).))----)......(((.(((((......).)))))))........))))(((..---.(.((((((((-.......)))))))).)....))) ( -29.30, z-score = -1.80, R) >consensus UUGUACG_CUUACAGGGUAUUUAAAUCUCU____GCCUGCCGAAAUGAACUCCUCCGCCUCGUUCAUUUUUCUGCGUGACCA___UGUCGGGUCAC_AAACCACGUGACCCAUCUUUUGUC ..............((((....................((.((((.((((...........))))...)))).))((((((.........))))))...........)))).......... (-18.72 = -18.60 + -0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:00:35 2011