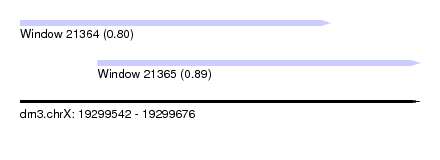
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,299,542 – 19,299,676 |
| Length | 134 |
| Max. P | 0.891679 |

| Location | 19,299,542 – 19,299,646 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.54 |
| Shannon entropy | 0.22509 |
| G+C content | 0.53951 |
| Mean single sequence MFE | -35.74 |
| Consensus MFE | -29.78 |
| Energy contribution | -30.14 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.796696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
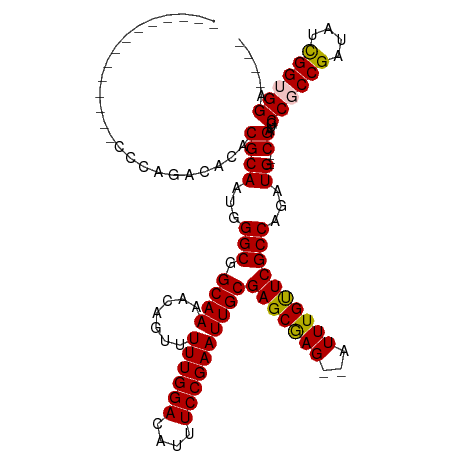
>dm3.chrX 19299542 104 + 22422827 --------------CCCAGACACACGCAUUGGGCGGCAAAACAGUUUUUGGACAUUUCCGAAUUGCGAGCAAG--AUUUGUUCGCCAGAUGCUGCGAUGCCGCCGAUAUCGGUGGAUGGA --------------.(((..(((.((...(.(((((((...((((.((((((....))))))..(((((((..--...))))))).....))))...))))))).)...)))))..))). ( -43.30, z-score = -3.87, R) >droSim1.chrX 14917072 97 + 17042790 --------------CCCAGACGCACGCAAUGGGCGGCAAAACAGUUUUUGGACAUUUCCGAAUUGCGAGCGAG--AUUUGUUCGCCAGAUG---CGAUGCCGCCGAUAUCGGUGGA---- --------------......(((.((..((.(((((((........((((((....))))))..((((((((.--..))))))))......---...))))))).))..)))))..---- ( -33.90, z-score = -1.36, R) >droSec1.super_8 1591607 97 + 3762037 --------------CCCAGACGCACGCAAUGGGCGGCAAAACAGUUUUUGGACAUUUCCGAAUUGCGAGCGAG--AUUUGCUCGCCAGAUG---CGAUGCCGCCGAUAUCGGUGGA---- --------------......(((.((..((.(((((((........((((((....))))))..((((((((.--..))))))))......---...))))))).))..)))))..---- ( -36.30, z-score = -1.91, R) >droYak2.chrX 9741090 113 - 21770863 GCUGGAGAGCACACGCAGGACACACGCAAUGGGCUGCAAAACAGUUUUUGGACAUUUCCGAAUUGCGAGCGAGAGAUUUGUUCGCCAGAUG---CGAUGCCGCCGAUAUUGGGGGU---- .((((.((((((.(((.(....).((((((((((((.....))))))(((((....))))))))))).))).).....))))).))))...---....(((.(((....))).)))---- ( -33.90, z-score = -0.28, R) >droEre2.scaffold_4690 9549575 97 + 18748788 -------------CCCAAG-CACACGCAAUGGGCUGCAAAACAGUUUUUGGACAUUUCCGAAUUGCGAGCGAG--AUUUGUUCGCCAGAUG---CGAUGCCGCCGAUAUUGGGGGU---- -------------((((((-(....)).((.(((.(((........((((((....))))))..((((((((.--..))))))))......---...))).))).)).)))))...---- ( -31.30, z-score = -0.86, R) >consensus ______________CCCAGACACACGCAAUGGGCGGCAAAACAGUUUUUGGACAUUUCCGAAUUGCGAGCGAG__AUUUGUUCGCCAGAUG___CGAUGCCGCCGAUAUCGGUGGA____ ....................(((.((..((.(((((((........((((((....))))))..(((((((((...)))))))))............))))))).))..)))))...... (-29.78 = -30.14 + 0.36)


| Location | 19,299,568 – 19,299,676 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 69.71 |
| Shannon entropy | 0.54339 |
| G+C content | 0.52111 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -20.96 |
| Energy contribution | -22.13 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.891679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
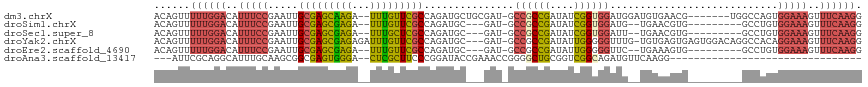
>dm3.chrX 19299568 108 + 22422827 ACAGUUUUUGGACAUUUCCGAAUUGCGAGCAAGA--UUUGUUCGCCAGAUGCUGCGAU-GCCGCCGAUAUCGGUGGAUGGAUGUGAACG-------UGGCCAGUGGAAAGUUUCAAGG ......((((((..((((((....(((((((...--..))))))).....((..(...-.((((((....))))))......(....))-------..))...))))))..)))))). ( -34.60, z-score = -1.17, R) >droSim1.chrX 14917098 101 + 17042790 ACAGUUUUUGGACAUUUCCGAAUUGCGAGCGAGA--UUUGUUCGCCAGAUGC---GAU-GCCGCCGAUAUCGGUGGAUG--UGAACGUG---------GCCUGUGGAAAGUUUCAAGG ......((((((..((((((....((((((((..--.))))))))(((..((---...-.((((((....))))))(((--....))).---------)))))))))))..)))))). ( -32.80, z-score = -1.35, R) >droSec1.super_8 1591633 101 + 3762037 ACAGUUUUUGGACAUUUCCGAAUUGCGAGCGAGA--UUUGCUCGCCAGAUGC---GAU-GCCGCCGAUAUCGGUGGAUU--UGAACGUG---------GCCUGUGGAAAGUUUCAAGG ......((((((..((((((....((((((((..--.))))))))(((..((---.((-(((((((....))))))...--....))).---------)))))))))))..)))))). ( -34.81, z-score = -1.93, R) >droYak2.chrX 9741130 113 - 21770863 ACAGUUUUUGGACAUUUCCGAAUUGCGAGCGAGAGAUUUGUUCGCCAGAUGC---GAU-GCCGCCGAUAUUGGGGGUUUG-UGUGAGUGAGUGGACAGGCCACAGGAAAGUUUCAAGG ......((((((..(((((.....(((((((((...)))))))))...((((---((.-(((.(((....))).))))))-)))......((((.....)))).)))))..)))))). ( -34.10, z-score = -1.10, R) >droEre2.scaffold_4690 9549601 101 + 18748788 ACAGUUUUUGGACAUUUCCGAAUUGCGAGCGAGA--UUUGUUCGCCAGAUGC---GAU-GCCGCCGAUAUUGGGGGUUC--UGAAAGUG---------GCCUGUGGAAAGUUUCAAGG ......((((((..((((((......((((((..--.))))))((((....(---(..-(((.(((....))).)))..--))....))---------))...))))))..)))))). ( -28.00, z-score = 0.07, R) >droAna3.scaffold_13417 4288931 81 - 6960332 ---AUUCGCAGGCAUUUGCAAGCGGCGAGUGGGA--CUCGCUUCCCGGAUACCGAAACCGGGGCUGCGGUCGGCAGAUGUUCAAGG-------------------------------- ---.......(((((((((..((((((((.....--)))....(((((.........)))))))))).....))))))))).....-------------------------------- ( -30.60, z-score = -0.76, R) >consensus ACAGUUUUUGGACAUUUCCGAAUUGCGAGCGAGA__UUUGUUCGCCAGAUGC___GAU_GCCGCCGAUAUCGGUGGAUG__UGAAAGUG_________GCCUGUGGAAAGUUUCAAGG ......((((((..(((((.....(((((((((...)))))))))...............((((((....))))))............................)))))..)))))). (-20.96 = -22.13 + 1.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:00:33 2011