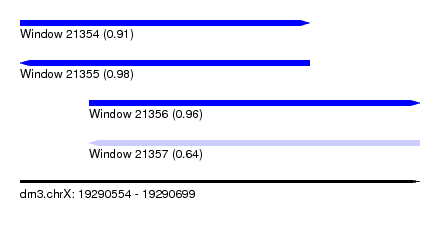
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,290,554 – 19,290,699 |
| Length | 145 |
| Max. P | 0.979925 |

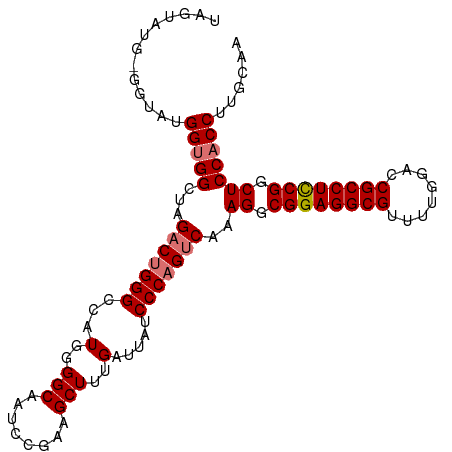
| Location | 19,290,554 – 19,290,659 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 91.65 |
| Shannon entropy | 0.13723 |
| G+C content | 0.56051 |
| Mean single sequence MFE | -43.03 |
| Consensus MFE | -38.54 |
| Energy contribution | -39.10 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19290554 105 + 22422827 UAGUAUG-GGUAUGGUGGCUAGCCUGGGCCAUGGGGCAAUACGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUUGCAA ..(((.(-(((...((((((......))))))(((((.........((((((((.....)))))))).((((((((........)))))))))))))))))))).. ( -45.70, z-score = -2.18, R) >droSim1.chrX 14911277 106 + 17042790 UAGUAUUUGGUAUGGUGGCUAGACUGGGCCAUGGGGCAAUCCGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUUGCAA .........(((.(((((...(((((((...(..(((........)))..).....)))))))..((.((((((((........)))))))).))))))).))).. ( -46.00, z-score = -2.70, R) >droSec1.super_8 1582669 104 + 3762037 UAGUAUG-G-UAUGGCGGCUAGACUGGGCCAUGGGGCAAUCCGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUUGCAA ..(((.(-(-(.....((((......))))..(((((.........((((((((.....)))))))).((((((((........)))))))))))))))).))).. ( -43.60, z-score = -1.96, R) >droEre2.scaffold_4690 9541064 99 + 18748788 -------UGAUGUGGUGGCUAGACGGGGCUAUGGGGCAAUCCGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUUCGGCUCCACCUUGCAA -------...((((((((...(((((((..(((((((........))))).))..)))).)))..((.((((((((........)))))))).)))))))..))). ( -36.80, z-score = -0.71, R) >consensus UAGUAUG_GGUAUGGUGGCUAGACUGGGCCAUGGGGCAAUCCGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUUGCAA .............(((((...(((((((...(..(((........)))..).....)))))))..((.((((((((........)))))))).)))))))...... (-38.54 = -39.10 + 0.56)

| Location | 19,290,554 – 19,290,659 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 91.65 |
| Shannon entropy | 0.13723 |
| G+C content | 0.56051 |
| Mean single sequence MFE | -38.48 |
| Consensus MFE | -36.50 |
| Energy contribution | -37.06 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19290554 105 - 22422827 UUGCAAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGUAUUGCCCCAUGGCCCAGGCUAGCCACCAUACC-CAUACUA ......((((((((((((((((........)))))))(((......((((.(((((........).)))).)))).)))...))))..))))).....-....... ( -39.50, z-score = -2.07, R) >droSim1.chrX 14911277 106 - 17042790 UUGCAAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGGAUUGCCCCAUGGCCCAGUCUAGCCACCAUACCAAAUACUA ......(((((((.((((((((........)))))))).))..(((((((..........(....)............)))))))...)))))............. ( -40.95, z-score = -2.82, R) >droSec1.super_8 1582669 104 - 3762037 UUGCAAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGGAUUGCCCCAUGGCCCAGUCUAGCCGCCAUA-C-CAUACUA ......(((((((.((((((((........)))))))).))..(((((((..........(....)............)))))))...)))))...-.-....... ( -40.55, z-score = -2.34, R) >droEre2.scaffold_4690 9541064 99 - 18748788 UUGCAAGGUGGAGCCGAAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGGAUUGCCCCAUAGCCCCGUCUAGCCACCACAUCA------- ..((.((..((.(((((((((((............)))))))))..((((.(((((.........))))).))))..)).))..)).))..........------- ( -32.90, z-score = -1.50, R) >consensus UUGCAAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGGAUUGCCCCAUGGCCCAGUCUAGCCACCAUACC_CAUACUA ......(((((((.((((((((........)))))))).))..(((((((.........((........)).......)))))))...)))))............. (-36.50 = -37.06 + 0.56)

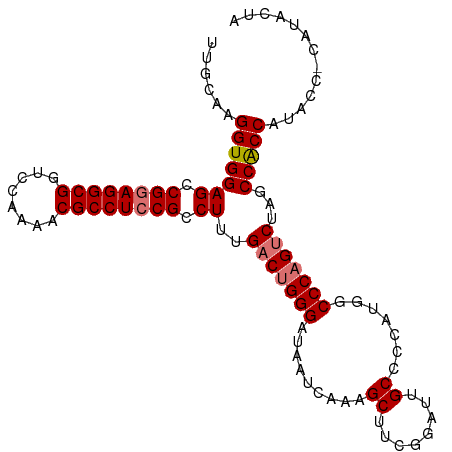
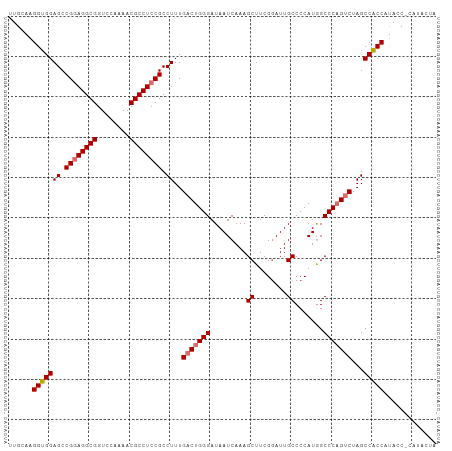
| Location | 19,290,579 – 19,290,699 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.30 |
| Shannon entropy | 0.09733 |
| G+C content | 0.57816 |
| Mean single sequence MFE | -47.98 |
| Consensus MFE | -44.14 |
| Energy contribution | -44.02 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.955106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19290579 120 + 22422827 GGCCAUGGGGCAAUACGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUUGCAAGCCAAAGUGGGCGGUGUUACUUUGCUUUCCCCAUACCCAG ((..((((((.((..(((((((((((((.....)))))))).((((((((........))))))))((.((((.(((.....))).)))))).......)))))..))))))))..)).. ( -48.50, z-score = -2.44, R) >droSim1.chrX 14911303 120 + 17042790 GGCCAUGGGGCAAUCCGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUUGCAAGCCAAAGUGGGCGGUGUUACUUUGCUUUCCCCAUCCCCAG ((..((((((.((..(((((((((((((.....)))))))).((((((((........))))))))((.((((.(((.....))).)))))).......)))))..))))))))..)).. ( -48.50, z-score = -2.12, R) >droSec1.super_8 1582693 120 + 3762037 GGCCAUGGGGCAAUCCGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUUGCAAGCCAAGGUGGGCGGUGUUACUUUGCUUUCCCCAUCCCCAG ((..((((((.((..(((((((((((((.....)))))))).((((((((........))))))))((.((((((((.....)))))))))).......)))))..))))))))..)).. ( -55.00, z-score = -3.45, R) >droYak2.chrX 9732108 114 - 21770863 GGCCAUGGGGCAAUCCGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUAGCAAGCCAAAGUGGGCGGUGUUACUUCGCUUUCCCCAG------ .....(((((.((..(((((((((((((.....)))))))).((((((((........))))))))........(((((.(((......)))..))))))))))..))))))).------ ( -45.90, z-score = -1.89, R) >droEre2.scaffold_4690 9541083 114 + 18748788 GGCUAUGGGGCAAUCCGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUUCGGCUCCACCUUGCAAGCCAAAGUGGGCGGUGUUACUUUGCUUUCCCCAG------ .....(((((.((..(((((((((((((.....)))))))).((((((((........))))))))((.((((.(((.....))).)))))).......)))))..))))))).------ ( -42.00, z-score = -0.97, R) >consensus GGCCAUGGGGCAAUCCGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUUGCAAGCCAAAGUGGGCGGUGUUACUUUGCUUUCCCCAU_CCCAG .....(((((.((..(((((((((((((.....)))))))).((((((((........))))))))((.((((.(((.....))).)))))).......)))))..)))))))....... (-44.14 = -44.02 + -0.12)

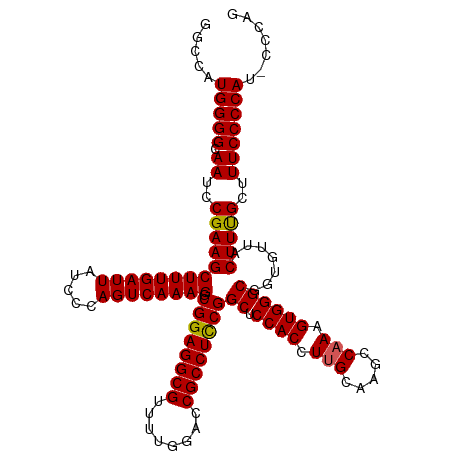
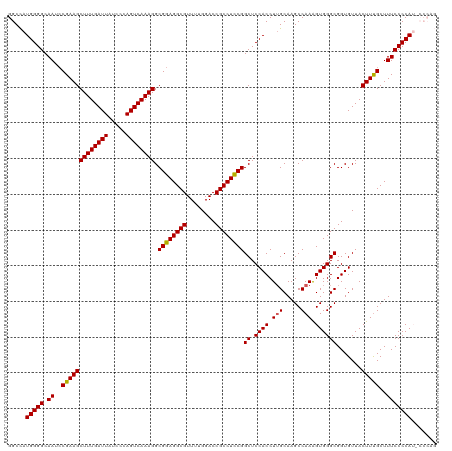
| Location | 19,290,579 – 19,290,699 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.30 |
| Shannon entropy | 0.09733 |
| G+C content | 0.57816 |
| Mean single sequence MFE | -46.56 |
| Consensus MFE | -41.88 |
| Energy contribution | -42.56 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.635026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19290579 120 - 22422827 CUGGGUAUGGGGAAAGCAAAGUAACACCGCCCACUUUGGCUUGCAAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGUAUUGCCCCAUGGCC ..((.(((((((((.((.((((......((((((((((.....)))))))).))((((((((........))))))))(((......))).........)))).)).)).))))))).)) ( -50.60, z-score = -2.46, R) >droSim1.chrX 14911303 120 - 17042790 CUGGGGAUGGGGAAAGCAAAGUAACACCGCCCACUUUGGCUUGCAAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGGAUUGCCCCAUGGCC ..((..((((((..(((((((.......((((((((((.....)))))))).))((((((((........)))))))).))))).))....(((((.........)))))))))))..)) ( -47.10, z-score = -1.01, R) >droSec1.super_8 1582693 120 - 3762037 CUGGGGAUGGGGAAAGCAAAGUAACACCGCCCACCUUGGCUUGCAAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGGAUUGCCCCAUGGCC ..((..((((((..(((((((.......((((((((((.....)))))))).))((((((((........)))))))).))))).))....(((((.........)))))))))))..)) ( -49.80, z-score = -1.54, R) >droYak2.chrX 9732108 114 + 21770863 ------CUGGGGAAAGCGAAGUAACACCGCCCACUUUGGCUUGCUAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGGAUUGCCCCAUGGCC ------.(((((.((.((((((......(((......)))...((((..((((.((((((((........)))))))).))))..))))..........))))))..)).)))))..... ( -46.10, z-score = -1.65, R) >droEre2.scaffold_4690 9541083 114 - 18748788 ------CUGGGGAAAGCAAAGUAACACCGCCCACUUUGGCUUGCAAGGUGGAGCCGAAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGGAUUGCCCCAUAGCC ------.(((((..............((((((((((((.....)))))))).))(((((((((............)))))))))...))..(((((.........))))))))))..... ( -39.20, z-score = -0.70, R) >consensus CUGGG_AUGGGGAAAGCAAAGUAACACCGCCCACUUUGGCUUGCAAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGGAUUGCCCCAUGGCC ......((((((..(((((((.......((((((((((.....)))))))).))((((((((........)))))))).))))).))....(((((.........))))))))))).... (-41.88 = -42.56 + 0.68)

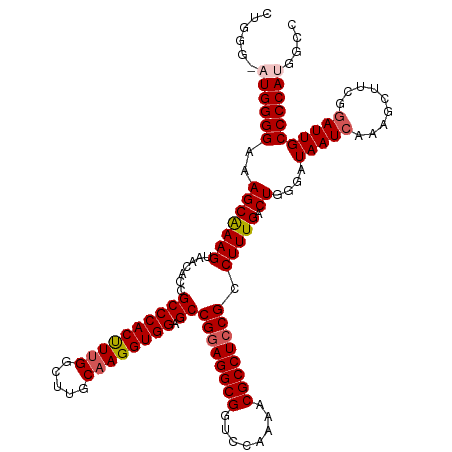
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:00:26 2011