
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,268,249 – 19,268,359 |
| Length | 110 |
| Max. P | 0.989555 |

| Location | 19,268,249 – 19,268,359 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.96 |
| Shannon entropy | 0.38337 |
| G+C content | 0.36242 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -14.55 |
| Energy contribution | -17.05 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.986272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
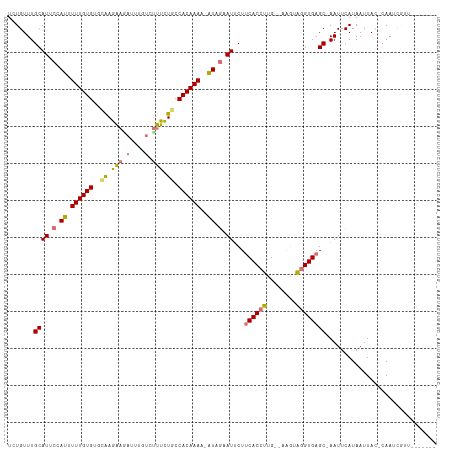
>dm3.chrX 19268249 110 + 22422827 UCUGUUUGCAUACCAUUUUUGUGUGCAAGAAGAUUUGUCUUUCUGCCACAAACUGUGGAAUGCUACACCUUGUUUAGUAGGUGAGC-AAUUCAUAAUUAC-CAAACGUUGGU---- ...(((((.........((((((.(((..((((....))))..))))))))).((((((.((((.(((((........))))))))-).)))))).....-)))))......---- ( -35.00, z-score = -3.48, R) >droSec1.super_8 1560659 108 + 3762037 UCUGUUUGCAUUCCAUUUUUGUGUGCGAGAAGACUUGUCCUUCUGCCACAAACUGUGGAAUUCUUCACCUUGUUUAGUGUGUGAGCCAAUUCAUAAUUAC-CAAUCGUU------- ...(((..((((((((.((((((.((.(((((.......)))))))))))))..)))))).....(((........)))))..)))..............-........------- ( -26.20, z-score = -2.26, R) >droYak2.chrX 9709619 105 - 21770863 UCUGUUUGCAUUCCAUUUUUGUGCAUGAAGAUUUCUGUCUCUUUGCCACAAAA-AUAGCAUUCUUCACCUUG--AAUUAGGUGAGCAAAUUCAUAAUUAC-UAACCUUU------- ...((((((.....(((((((((((...((((....))))...)).)))))))-))........((((((..--....))))))))))))..........-........------- ( -25.50, z-score = -3.45, R) >droEre2.scaffold_4690 9519803 110 + 18748788 UCUGUUUGCAUGCCAUUUUUGUG--CAAGAAAAUUUGUCUUUCUACCACAAAA-AUAACAUUCUUCACCUUG--AGGUAGGUGAGC-AAUUCAUAAUUACAUAAUUACUUCCCCUU .....(((((((..(((((((((--..(((((.......)))))..)))))))-))..)))...((((((..--....))))))))-))........................... ( -22.70, z-score = -2.47, R) >consensus UCUGUUUGCAUUCCAUUUUUGUGUGCAAGAAGAUUUGUCUUUCUGCCACAAAA_AUAGAAUUCUUCACCUUG__AAGUAGGUGAGC_AAUUCAUAAUUAC_CAAUCGUU_______ ...(((.((((((((((((((((.(((.(((((....))))).)))))))))).)))))))...((((((........)))))))).))).......................... (-14.55 = -17.05 + 2.50)


| Location | 19,268,249 – 19,268,359 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.96 |
| Shannon entropy | 0.38337 |
| G+C content | 0.36242 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -16.41 |
| Energy contribution | -17.48 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
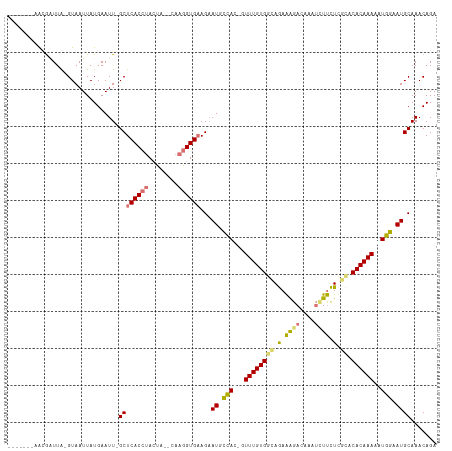
>dm3.chrX 19268249 110 - 22422827 ----ACCAACGUUUG-GUAAUUAUGAAUU-GCUCACCUACUAAACAAGGUGUAGCAUUCCACAGUUUGUGGCAGAAAGACAAAUCUUCUUGCACACAAAAAUGGUAUGCAAACAGA ----......(((((-............(-((((((((........))))).))))..(((...(((((((((..((((....))))..))).))))))..)))....)))))... ( -31.80, z-score = -3.09, R) >droSec1.super_8 1560659 108 - 3762037 -------AACGAUUG-GUAAUUAUGAAUUGGCUCACACACUAAACAAGGUGAAGAAUUCCACAGUUUGUGGCAGAAGGACAAGUCUUCUCGCACACAAAAAUGGAAUGCAAACAGA -------..(((((.-((....)).)))))((.....((((......))))....((((((...((((((((((((((.....)))))).)).))))))..))))))))....... ( -28.80, z-score = -2.66, R) >droYak2.chrX 9709619 105 + 21770863 -------AAAGGUUA-GUAAUUAUGAAUUUGCUCACCUAAUU--CAAGGUGAAGAAUGCUAU-UUUUGUGGCAAAGAGACAGAAAUCUUCAUGCACAAAAAUGGAAUGCAAACAGA -------....(((.-(((.......((((..((((((....--..)))))).)))).((((-(((((((.((..((((......))))..)))))))))))))..))).)))... ( -27.50, z-score = -2.85, R) >droEre2.scaffold_4690 9519803 110 - 18748788 AAGGGGAAGUAAUUAUGUAAUUAUGAAUU-GCUCACCUACCU--CAAGGUGAAGAAUGUUAU-UUUUGUGGUAGAAAGACAAAUUUUCUUG--CACAAAAAUGGCAUGCAAACAGA ..(((..((((((((((....)))).)))-)))..)))(((.--...)))...(.(((((((-(((((((..((((((.....))))))..--)))))))))))))).)....... ( -30.20, z-score = -3.08, R) >consensus _______AACGAUUA_GUAAUUAUGAAUU_GCUCACCUACUA__CAAGGUGAAGAAUGCCAC_GUUUGUGGCAGAAAGACAAAUCUUCUCGCACACAAAAAUGGAAUGCAAACAGA ..............................((((((((........))))))...((((((...((((((((((.((((....)))).)))).))))))..))))))))....... (-16.41 = -17.48 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:00:16 2011