
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,259,556 – 19,259,669 |
| Length | 113 |
| Max. P | 0.645208 |

| Location | 19,259,556 – 19,259,669 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 63.27 |
| Shannon entropy | 0.68067 |
| G+C content | 0.46157 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -11.38 |
| Energy contribution | -12.75 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.645208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19259556 113 + 22422827 CGCGGUUUUCCUUUUAAUUUU-GCAUCCUUGUUUUGGGAAAAAUGUACGCCUUUUAACGAUAGGCGCUU-----UGCCCCUCUGAGAGAACCAUUUUGUAGGGGCCAAAUCUCCUACUA ...(((((((..........(-(((((((......))))....))))(((((.........)))))...-----...........))))))).....(((((((......))))))).. ( -28.80, z-score = -1.02, R) >droSim1.chrX 14884378 117 + 17042790 CGCUGUCUUCCUUUUAAUUUU-GCAUCCUUGUUUUGGGGAAGUGGUACGU-UUUUAACGAUAGGCGCUUUGUGCUGCCCCUCCAAGAGCACCAUUUUGCAGGGGCCAAGUCUCCUGCCA .((((.(((((((..(((...-........)))..))))))))))).(((-.....)))...(((((.....)).((((((.(((((......))))).))))))..........))). ( -33.80, z-score = -0.31, R) >droSec1.super_8 1552086 117 + 3762037 CGCUGUUUUCCUUUUAAUUUU-GCAUCCUUGUUUUGGGGAAGUGGUACGU-UUUUAACGAUAGGCGCUUUGUGCUGCCCCUCCAAGAGCACCAUUUUGCAGGGGACAAGUCUCCUGCUA .((..(((((((.........-(((....)))...)))))))..))..((-((((...((..(((((.....)).)))..)).))))))........((((((((....)))))))).. ( -34.60, z-score = -1.02, R) >droYak2.chrX 9700766 107 - 21770863 CGCUGUUUUCCCUUUAUUUUUUGCAUCCUUGUUU-GGGGAAAAUAUACGU-UUUUAACGAUGGGCGUUUC-----GCCCCUCUAAGAGUGCCAUUUUGUAGUAGUCUCCUACUA----- (((((((((((((.........(((....)))..-)))))))))......-.......((.(((((...)-----)))).))....))))........((((((....))))))----- ( -26.10, z-score = -2.06, R) >droEre2.scaffold_4690 9511596 98 + 18748788 CGCUGUUUUCCUCUUAUUUUUUGCAUCCUUGUUU-GGGGCAAAUGUACGU-UUUUAACGAUGGGCGUUU------GCCCCUCUAAAAGUGCCAUUUUGUAGGGGCC------------- ..........(((((((.....((((..(((...-((((((((((..(((-(......))))..)))))------)))))..)))..))))......)))))))..------------- ( -31.60, z-score = -2.75, R) >droWil1.scaffold_181150 1556524 102 - 4952429 CGCCAUAGCCUCUCAAACUCCUAGAGACCUA----GAGACGUGUUCGCCUCUUCUGUGGUUGUCAGCUU------GCCAUCAAAAACCAGUCAAAGAACUGGGCUCCUCAUC------- .(((((((..((((.........))))...(----(((.((....)).)))).)))))))((..(((((------(....)))...(((((......))))))))...))..------- ( -21.40, z-score = 0.28, R) >consensus CGCUGUUUUCCUUUUAAUUUU_GCAUCCUUGUUU_GGGGAAAAUGUACGU_UUUUAACGAUAGGCGCUU______GCCCCUCUAAGAGAACCAUUUUGCAGGGGCCAACUCUCC_____ ((((.(((((((.......................))))))).....((........))...)))).........((((((.(((((......))))).)))))).............. (-11.38 = -12.75 + 1.37)

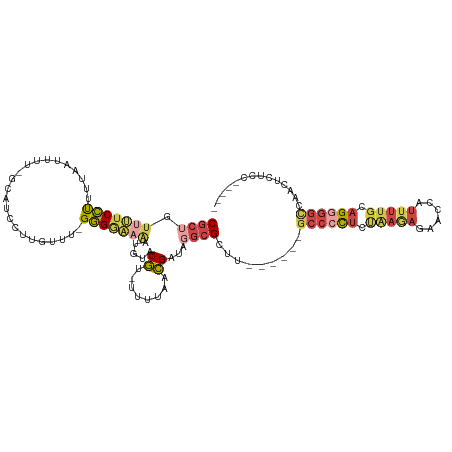
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:00:13 2011