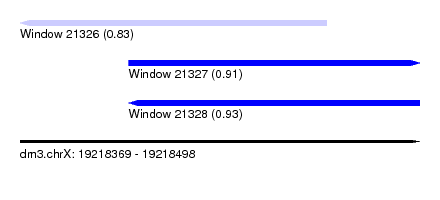
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,218,369 – 19,218,498 |
| Length | 129 |
| Max. P | 0.932129 |

| Location | 19,218,369 – 19,218,468 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 54.70 |
| Shannon entropy | 0.72542 |
| G+C content | 0.44621 |
| Mean single sequence MFE | -23.48 |
| Consensus MFE | -7.59 |
| Energy contribution | -8.28 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19218369 99 - 22422827 AGCGAAUUUUUGCCUUCCAAUGCUGGUGU-GAUGUC-UCAUCGCCAUCCCCAUCAUCAUCAUUGCCAUCGUAGUGAAUGAAAUGAUGAU----GUAUUCGAUUUC- .((((....))))..((.(((((((((((-((....-))).)))))....(((((((((((((((....)))))).))))..)))))..----))))).))....- ( -25.00, z-score = -1.48, R) >droEre2.scaffold_4690 9475139 76 - 18748788 AGCAAAU---UGCCUUCCUAUGCUG--GC-GAUGUC-CCAUCAGCGCCAUCGUGUU------------------GAAUGAAACGAUAAU----GUAUUCAAUAUC- .....((---((((..........)--))-)))((.-.(((((((((....)))))------------------).)))..))((((.(----(....)).))))- ( -16.60, z-score = -1.02, R) >droSim1.chrX 14858114 79 - 17042790 AGCGCAUUUUUGCCUUCCUAUGCUG--GU-GAUGUC-UCAUCCUCAUCACCAUCAUC------------------AAUGAAAUGAUAAU----GUAUUCGAUUUC- ((.(((....)))))((..((((((--((-((((..-.......))))))))..(((------------------(......))))...----))))..))....- ( -16.30, z-score = -1.45, R) >droGri2.scaffold_15081 1610112 103 - 4274704 AGCUCAUUUCAGC--GACGACGGCGACGUCAACGUCAACGUCAACGUCGAC-UGAGAGAGCGGGCAGCAAGCACACAUCACAUGUUGACCUUGAUAUUCGCAUUCG .((((.(((((((--(((((((..((((....))))..))))...)))).)-)))))))))((((((((.(.........).))))).)))............... ( -36.00, z-score = -2.64, R) >consensus AGCGAAUUUUUGCCUUCCUAUGCUG__GU_GAUGUC_UCAUCAACAUCACCAUCAUC_________________GAAUGAAAUGAUAAU____GUAUUCGAUUUC_ .(((......)))......(((.((((((.((((....)))).)))))).)))..................................................... ( -7.59 = -8.28 + 0.69)

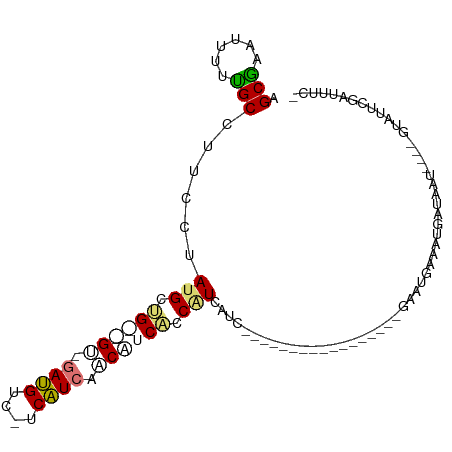
| Location | 19,218,404 – 19,218,498 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 53.68 |
| Shannon entropy | 0.70564 |
| G+C content | 0.50680 |
| Mean single sequence MFE | -27.59 |
| Consensus MFE | -10.10 |
| Energy contribution | -9.60 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.909570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19218404 94 + 22422827 -----GGCAAUGAUGAUGAUGGGGAUGGCGAUGA-GACAUCACACCAGCAUUGGAAGGCAAAAAUUCGCUGUGGGCCAUUGACUGGCUAACUUUUGCCAG---------- -----(((((....((((.(((.((((.(.....-).))))...))).))))....(((........)))((.(((((.....))))).))..)))))..---------- ( -26.40, z-score = -0.64, R) >droEre2.scaffold_4690 9475174 71 + 18748788 -----------------------GGCGCUGAUGG-GACAUCG--CCAGCAUAGGAAGGCAA---UUUGCUGUGGGCCAAUGACUGGCUAACUUUGGCCAG---------- -----------------------(((((((.(((-....)))--.))))((((.(((....---))).))))..))).....(((((((....)))))))---------- ( -24.20, z-score = -0.79, R) >droSim1.chrX 14858139 84 + 17042790 -------------UGAUGAUGGUGAUGAGGAUGA-GACAUCA--CCAGCAUAGGAAGGCAAAAAUGCGCUGUGGGCCAAUGACUGGCUAACUUUUGCCAG---------- -------------..(((.((((((((.......-..)))))--))).))).....(((((((.......((.(((((.....))))).)))))))))..---------- ( -27.31, z-score = -2.55, R) >droGri2.scaffold_15081 1610152 103 + 4274704 UGCCCGCUCUCUCAGUCGACGUUGACGUUGACGUUGACGUCG--CCGUCGUCG-----CUGAAAUGAGCUCUAAGAAAAUAAUUAAAUAGCAGCAGUCGGCAAACAAAAA ((((.(((...((((((((((.(((((((......)))))))--.)))))..)-----)))).....((((((..............))).)))))).))))........ ( -32.44, z-score = -2.40, R) >consensus _____________UGAUGAUGGUGACGAUGAUGA_GACAUCA__CCAGCAUAGGAAGGCAAAAAUGCGCUGUGGGCCAAUGACUGGCUAACUUUUGCCAG__________ .............................((((....))))......((.......(((........)))....))......(((((........))))).......... (-10.10 = -9.60 + -0.50)

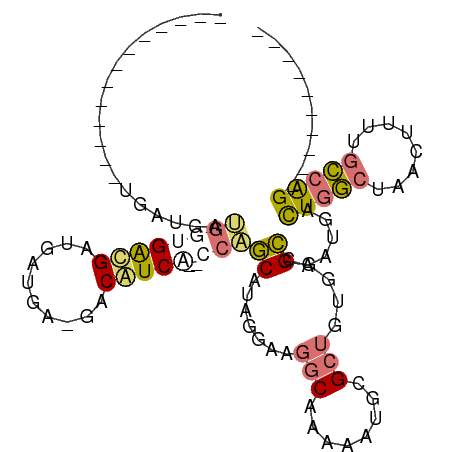
| Location | 19,218,404 – 19,218,498 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 53.68 |
| Shannon entropy | 0.70564 |
| G+C content | 0.50680 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -9.00 |
| Energy contribution | -9.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19218404 94 - 22422827 ----------CUGGCAAAAGUUAGCCAGUCAAUGGCCCACAGCGAAUUUUUGCCUUCCAAUGCUGGUGUGAUGUC-UCAUCGCCAUCCCCAUCAUCAUCAUUGCC----- ----------..((((((((((.((((.....))))........))))))))))...(((((.(((((.((((.(-.....).)))).....))))).)))))..----- ( -25.50, z-score = -1.75, R) >droEre2.scaffold_4690 9475174 71 - 18748788 ----------CUGGCCAAAGUUAGCCAGUCAUUGGCCCACAGCAAA---UUGCCUUCCUAUGCUGG--CGAUGUC-CCAUCAGCGCC----------------------- ----------..((((.......(((((.(((.((......((...---..))...)).)))))))--)((((..-.)))).).)))----------------------- ( -18.90, z-score = -0.22, R) >droSim1.chrX 14858139 84 - 17042790 ----------CUGGCAAAAGUUAGCCAGUCAUUGGCCCACAGCGCAUUUUUGCCUUCCUAUGCUGG--UGAUGUC-UCAUCCUCAUCACCAUCAUCA------------- ----------..(((((((((..(((((...))))).........))))))))).....(((.(((--(((((..-.......)))))))).)))..------------- ( -23.90, z-score = -2.54, R) >droGri2.scaffold_15081 1610152 103 - 4274704 UUUUUGUUUGCCGACUGCUGCUAUUUAAUUAUUUUCUUAGAGCUCAUUUCAG-----CGACGACGG--CGACGUCAACGUCAACGUCAACGUCGACUGAGAGAGCGGGCA ........((((...........(((((........)))))((((.((((((-----((((((((.--.((((....))))..))))...)))).)))))))))).)))) ( -35.20, z-score = -2.70, R) >consensus __________CUGGCAAAAGUUAGCCAGUCAUUGGCCCACAGCGAAUUUUUGCCUUCCUAUGCUGG__CGAUGUC_UCAUCAACAUCACCAUCAUCA_____________ ..........(((((........((((.....)))).....(((......)))........)))))...((((....))))............................. ( -9.00 = -9.00 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 02:00:03 2011