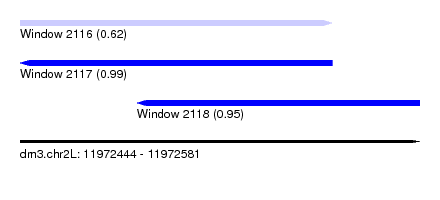
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,972,444 – 11,972,581 |
| Length | 137 |
| Max. P | 0.985373 |

| Location | 11,972,444 – 11,972,551 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 86.33 |
| Shannon entropy | 0.23807 |
| G+C content | 0.51577 |
| Mean single sequence MFE | -32.06 |
| Consensus MFE | -22.25 |
| Energy contribution | -23.77 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.623513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11972444 107 + 23011544 UGCUUAACGGAAGCGGUCGCAACCGUUAGACGCUGCAUCCAAAGCUAUAU-----GCAGCUAUAGCUGCAUCUGAAUCCCAGCUCUCGACGGCGAUCCCAACCAAAUGCAAU (((.....((..(.((((((...(((((((.((((.......((....((-----(((((....)))))))))......))))))).)))))))))).)..))....))).. ( -32.72, z-score = -2.04, R) >droSim1.chr2L 11779485 107 + 22036055 UGCUUAACGGAAGCGGUCGCAACCGUUAGACGCUGCAUCCAAAGCUAUAU-----GCAGCUAUAGCUGCAUCUGAAUCCCAACUCUUGCCGGCGAUCCCAACCAAAUGCAAU ((((((((((..((....))..))))))).(((((((...........((-----(((((....)))))))..((........)).)).))))).............))).. ( -29.70, z-score = -1.46, R) >droSec1.super_16 171775 107 + 1878335 UGCUUAACGGAAGCGGUCGCAACCGUUAGACGCUGCAUCCAAAGCUAUAU-----GCAGCUAUAGCUGCAUCUGAAUCCCAACUCUUGCCGGCGAUCCCAACCAAAUGCAAU ((((((((((..((....))..))))))).(((((((...........((-----(((((....)))))))..((........)).)).))))).............))).. ( -29.70, z-score = -1.46, R) >droYak2.chr2L 8394788 112 + 22324452 UGCUUAACGGAAGCGUUCGCCACCGUUAGACGCUGGAUCCAAAGCUAUACUCUAAGCAGCUAUAGCUGCAUCUGAAUCCCAGCUCUUGCCGGCGAUCCCAGCCAAAUGCAAU ((((((((((..((....))..)))))))..(((((...................(((((....)))))......(((((.((....)).)).))).))))).....))).. ( -35.40, z-score = -2.19, R) >droAna3.scaffold_12943 2768097 93 - 5039921 UGCUUAACGGAAGCGGUCGCAACCGUUAGACGCUGGAUCCGAAGCUAUA---------------GCUGAA----GAGCUGAGCUGUUGCCACCGAUCCCAGCCAAAUGCAAU ((((((((((..((....))..)))))))..((((((((.(..((.(((---------------(((.(.----....).)))))).))..).))).))))).....))).. ( -32.80, z-score = -2.62, R) >consensus UGCUUAACGGAAGCGGUCGCAACCGUUAGACGCUGCAUCCAAAGCUAUAU_____GCAGCUAUAGCUGCAUCUGAAUCCCAGCUCUUGCCGGCGAUCCCAACCAAAUGCAAU ((((((((((..((....))..))))))).(((((....(((((((.........(((((....)))))...........)))).))).))))).............))).. (-22.25 = -23.77 + 1.52)

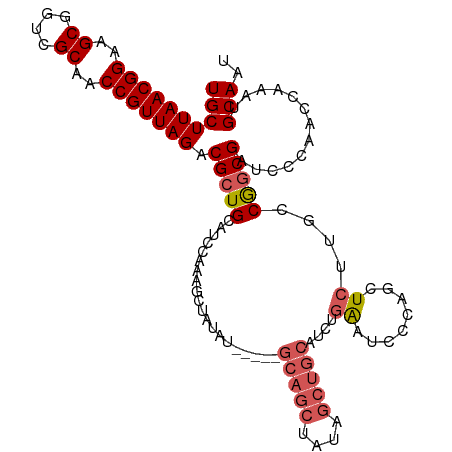
| Location | 11,972,444 – 11,972,551 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 86.33 |
| Shannon entropy | 0.23807 |
| G+C content | 0.51577 |
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -33.46 |
| Energy contribution | -35.30 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11972444 107 - 23011544 AUUGCAUUUGGUUGGGAUCGCCGUCGAGAGCUGGGAUUCAGAUGCAGCUAUAGCUGC-----AUAUAGCUUUGGAUGCAGCGUCUAACGGUUGCGACCGCUUCCGUUAAGCA .((((((((((((((((((.(((.(....).))))))))..(((((((....)))))-----)).)))))..)))))))).(..((((((..((....))..))))))..). ( -42.90, z-score = -2.56, R) >droSim1.chr2L 11779485 107 - 22036055 AUUGCAUUUGGUUGGGAUCGCCGGCAAGAGUUGGGAUUCAGAUGCAGCUAUAGCUGC-----AUAUAGCUUUGGAUGCAGCGUCUAACGGUUGCGACCGCUUCCGUUAAGCA .((((((((((((((((((.(((((....))))))))))..(((((((....)))))-----)).)))))..)))))))).(..((((((..((....))..))))))..). ( -44.30, z-score = -3.30, R) >droSec1.super_16 171775 107 - 1878335 AUUGCAUUUGGUUGGGAUCGCCGGCAAGAGUUGGGAUUCAGAUGCAGCUAUAGCUGC-----AUAUAGCUUUGGAUGCAGCGUCUAACGGUUGCGACCGCUUCCGUUAAGCA .((((((((((((((((((.(((((....))))))))))..(((((((....)))))-----)).)))))..)))))))).(..((((((..((....))..))))))..). ( -44.30, z-score = -3.30, R) >droYak2.chr2L 8394788 112 - 22324452 AUUGCAUUUGGCUGGGAUCGCCGGCAAGAGCUGGGAUUCAGAUGCAGCUAUAGCUGCUUAGAGUAUAGCUUUGGAUCCAGCGUCUAACGGUGGCGAACGCUUCCGUUAAGCA ..........(((((((((.(((((....)))))))))).((((((((....)))))((((((.....)))))))))))))(..((((((.(((....))).))))))..). ( -45.30, z-score = -2.48, R) >droAna3.scaffold_12943 2768097 93 + 5039921 AUUGCAUUUGGCUGGGAUCGGUGGCAACAGCUCAGCUC----UUCAGC---------------UAUAGCUUCGGAUCCAGCGUCUAACGGUUGCGACCGCUUCCGUUAAGCA ..........((((((.((((((....))(((.((((.----...)))---------------)..))).)))).))))))(..((((((..((....))..))))))..). ( -33.20, z-score = -1.71, R) >consensus AUUGCAUUUGGUUGGGAUCGCCGGCAAGAGCUGGGAUUCAGAUGCAGCUAUAGCUGC_____AUAUAGCUUUGGAUGCAGCGUCUAACGGUUGCGACCGCUUCCGUUAAGCA .((((((((((((((((((.(((((....))))))))))....(((((....)))))........)))))..)))))))).(..((((((..((....))..))))))..). (-33.46 = -35.30 + 1.84)

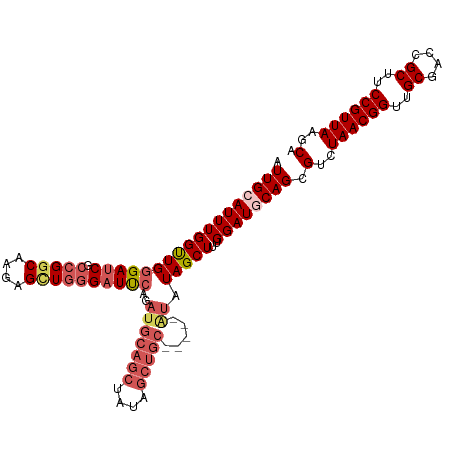
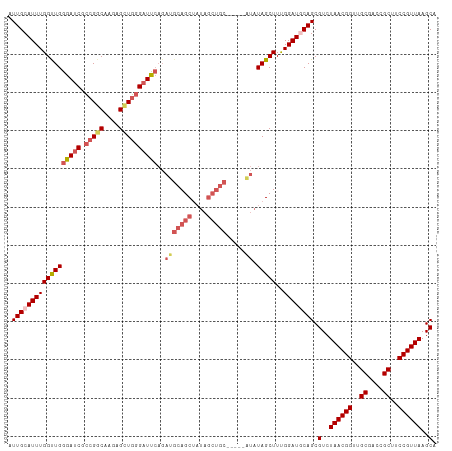
| Location | 11,972,484 – 11,972,581 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 85.40 |
| Shannon entropy | 0.23613 |
| G+C content | 0.48777 |
| Mean single sequence MFE | -36.55 |
| Consensus MFE | -31.55 |
| Energy contribution | -33.55 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11972484 97 - 23011544 AGCAGCAGCUGCAGUUUGUUGCUGCAACUAAUUGCAUUUGGUUGGGAUCGCCGUCGAGAGCUGGGAUUCAGAUGCAGCUAUAGCUGCAUAUAGCUUU .((((((((........))))))))((((((......)))))).(((((.(((.(....).))))))))..(((((((....)))))))........ ( -38.40, z-score = -2.05, R) >droSim1.chr2L 11779525 97 - 22036055 AGCAGCAGCUGCAGUUUGUUGCUGCAACUAAUUGCAUUUGGUUGGGAUCGCCGGCAAGAGUUGGGAUUCAGAUGCAGCUAUAGCUGCAUAUAGCUUU .((((((((........))))))))((((((......)))))).(((((.(((((....))))))))))..(((((((....)))))))........ ( -39.80, z-score = -2.74, R) >droSec1.super_16 171815 97 - 1878335 AGCAGCAGCUGCAGUUUGUUGCUGCAACUAAUUGCAUUUGGUUGGGAUCGCCGGCAAGAGUUGGGAUUCAGAUGCAGCUAUAGCUGCAUAUAGCUUU .((((((((........))))))))((((((......)))))).(((((.(((((....))))))))))..(((((((....)))))))........ ( -39.80, z-score = -2.74, R) >droAna3.scaffold_12943 2768137 83 + 5039921 GGCAGCAGCUGCAGUUUGUUGCUGCAACUAAUUGCAUUUGGCUGGGAUCGGUGGCAACAGCUCAGCUC----UUCAGCUAUAGCUUC---------- .((((((((........))))))))........((...((((((((...(.((((....)).)).)..----))))))))..))...---------- ( -28.20, z-score = -0.35, R) >consensus AGCAGCAGCUGCAGUUUGUUGCUGCAACUAAUUGCAUUUGGUUGGGAUCGCCGGCAAGAGCUGGGAUUCAGAUGCAGCUAUAGCUGCAUAUAGCUUU .((((((((........))))))))((((((......)))))).(((((.(((((....))))))))))..(((((((....)))))))........ (-31.55 = -33.55 + 2.00)

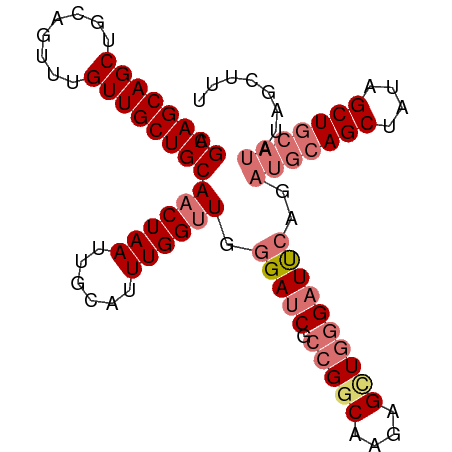
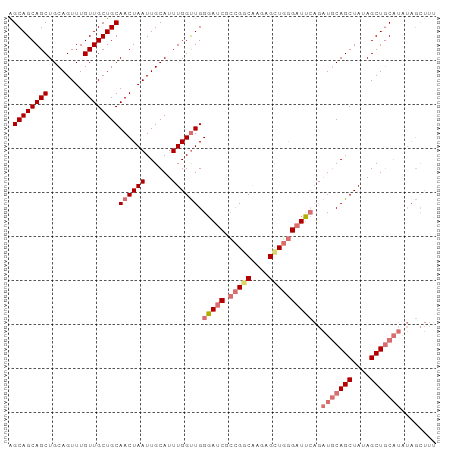
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:06 2011