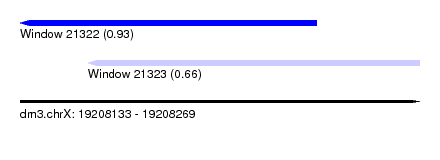
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,208,133 – 19,208,269 |
| Length | 136 |
| Max. P | 0.933386 |

| Location | 19,208,133 – 19,208,234 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.87 |
| Shannon entropy | 0.40320 |
| G+C content | 0.36390 |
| Mean single sequence MFE | -23.54 |
| Consensus MFE | -17.08 |
| Energy contribution | -18.68 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
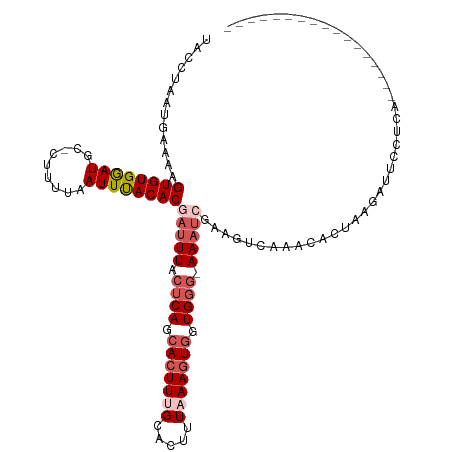
>dm3.chrX 19208133 101 - 22422827 UACCUAAUGAAAAGUGUGGAUGC-CUUUUAAUUUACACGAUUUACUCAGCAAUUUGCACUUUAAAGUGGUGGG-AAAUCGAAGUCAAGCACUAAGAUUCUUGA----------------- .............((((((((..-......))))))))(((((.((((.((.((((.....)))).)).))))-)))))....(((((..........)))))----------------- ( -19.70, z-score = -0.31, R) >droSim1.chrX 14847653 119 - 17042790 UACCUAAUGAAAAGUGUGGAUGUGCUUUUAAUUUACACGAUUUACUCAGCACUUUGCACUUUAAAGUGGUGGG-AAAUCGUAGUCAAACACUAAGAUUCCUCACGUUCUUUAAGUUCCCA .((.(((.(((..(((.(((....(((....((((((((((((.((((.(((((((.....))))))).))))-))))))).)).)))....)))..))).))).))).))).))..... ( -30.00, z-score = -2.32, R) >droSec1.super_8 1501472 119 - 3762037 UACCUAAUGAAAAGUGUGGAUGUGCUUUUAAUUUACACGAUUUACUCAGCACUUGGCACUUUAAAGUGGUGGG-AAAUCGAAGUCAAACACUAAGAUUCCUCACGUUCUUUAAGUUCUCA ........(((.(((((.((.(((((...((((.....)))).....))))))).)))))((((((..(((((-.((((..(((.....)))..)))).)))))...)))))).)))... ( -26.80, z-score = -1.13, R) >droYak2.chrX 9653498 102 + 21770863 UACCUAAUGAAAAGUGUGAAUGC-AUUUUAAUUCACACAAUUUACUCAGCACUUUGUACCUUAAAGUGGUGGGAAAACCGAAGUCAAACACCAAGAUUCCACA----------------- .......(((...((((((((..-......))))))))......((((.(((((((.....))))))).))))..........))).................----------------- ( -24.70, z-score = -2.57, R) >droEre2.scaffold_4690 9465690 97 - 18748788 UACCCAAUGAAAAGUGUGAAUGC-AUUUUAAUUUACACUAUUUACCCAGCACUUUGCACUUUUAAGUACUGC--AAUUC-----CUCACGUUAGCAGUUCCCAAA--------------- .......((((.(((((((((..-......))))))))).))))....((.....))........(.(((((--(((..-----.....))).))))).).....--------------- ( -16.50, z-score = -1.95, R) >consensus UACCUAAUGAAAAGUGUGGAUGC_CUUUUAAUUUACACGAUUUACUCAGCACUUUGCACUUUAAAGUGGUGGG_AAAUCGAAGUCAAACACUAAGAUUCCUCA_________________ .............((((((((.........))))))))(((((.((((.(((((((.....))))))).)))).)))))......................................... (-17.08 = -18.68 + 1.60)

| Location | 19,208,156 – 19,208,269 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.98 |
| Shannon entropy | 0.30284 |
| G+C content | 0.39571 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -18.58 |
| Energy contribution | -19.58 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
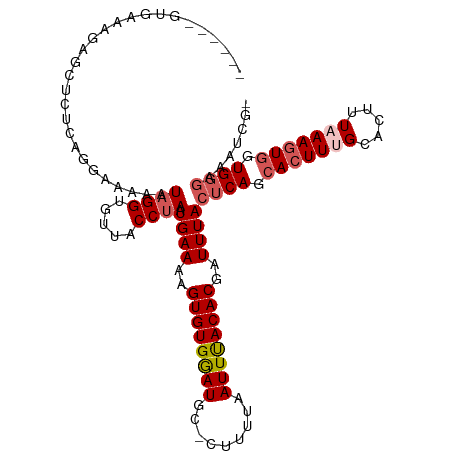
>dm3.chrX 19208156 113 - 22422827 CGCAUUGUGAGAGAGGUCUCAGGAAAAAUAGGUGUUACCUAAUGAAAAGUGUGGAUGC-CUUUUAAUUUACACGAUUUACUCAGCAAUUUGCACUUUAAAGUGGUGGGAAAUCG- ....((.(((((....))))).))....((((.....)))).......((((((((..-......))))))))(((((.((((.((.((((.....)))).)).))))))))).- ( -28.00, z-score = -1.41, R) >droSim1.chrX 14847693 108 - 17042790 ------GUGAGAGAGCUCUCGGGAAAAAUAGGUGUUACCUAAUGAAAAGUGUGGAUGUGCUUUUAAUUUACACGAUUUACUCAGCACUUUGCACUUUAAAGUGGUGGGAAAUCG- ------.(((((....))))).......((((.....)))).......((((((((.........))))))))(((((.((((.(((((((.....))))))).))))))))).- ( -31.70, z-score = -2.93, R) >droSec1.super_8 1501512 108 - 3762037 ------GUGACAGAGCUCUCGGGAAAAAUAGGUGUUACCUAAUGAAAAGUGUGGAUGUGCUUUUAAUUUACACGAUUUACUCAGCACUUGGCACUUUAAAGUGGUGGGAAAUCG- ------......................((((.....)))).......((((((((.........))))))))(((((.((((.(((((.........))))).))))))))).- ( -25.80, z-score = -0.84, R) >droYak2.chrX 9653521 108 + 21770863 -----AGUGAAAGAGUUCUCCGG-GGAAUAGGUGUUACCUAAUGAAAAGUGUGAAUGC-AUUUUAAUUCACACAAUUUACUCAGCACUUUGUACCUUAAAGUGGUGGGAAAACCG -----.........(((((....-))))).(((...............((((((((..-......))))))))......((((.(((((((.....))))))).))))...))). ( -30.40, z-score = -2.33, R) >droEre2.scaffold_4690 9465708 114 - 18748788 CGCGCUGGGAAAGAGUUCUCAGGAAAAAUAGGUGCUACCCAAUGAAAAGUGUGAAUGC-AUUUUAAUUUACACUAUUUACCCAGCACUUUGCACUUUUAAGUACUGCAAUUCCUC .((.(((((((....)))))))........((((((......((((.(((((((((..-......))))))))).))))....((.....)).......))))))))........ ( -28.10, z-score = -1.74, R) >consensus ______GUGAAAGAGCUCUCAGGAAAAAUAGGUGUUACCUAAUGAAAAGUGUGGAUGC_CUUUUAAUUUACACGAUUUACUCAGCACUUUGCACUUUAAAGUGGUGGGAAAUCG_ ............................((((.....)))).((((..((((((((.........))))))))..))))((((.(((((((.....))))))).))))....... (-18.58 = -19.58 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:59 2011