
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,172,574 – 19,172,637 |
| Length | 63 |
| Max. P | 0.999285 |

| Location | 19,172,574 – 19,172,637 |
|---|---|
| Length | 63 |
| Sequences | 3 |
| Columns | 68 |
| Reading direction | forward |
| Mean pairwise identity | 64.77 |
| Shannon entropy | 0.46525 |
| G+C content | 0.37970 |
| Mean single sequence MFE | -20.33 |
| Consensus MFE | -16.71 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999285 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
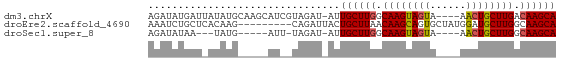
>dm3.chrX 19172574 63 + 22422827 AGAUAUGAUUAUAUGCAAGCAUCGUAGAU-AUUGCUUGGCAAGUAGUA----AACUGCUUGACAAGCA .(((((..(((((((....))).))))))-)))(((((.(((((((..----..))))))).))))). ( -18.80, z-score = -2.42, R) >droEre2.scaffold_4690 9430753 59 + 18748788 AAAUCUGCUCACAAG---------CAGAUUACUGCUUAACAAGCAGUGCUAUGGAUGCUUGGCAAGCA .((((((((....))---------))))))...((((..((((((...(....).))))))..)))). ( -22.80, z-score = -3.12, R) >droSec1.super_8 1467207 54 + 3762037 AGAUAUAA---UAUG-----AUU-UAGAU-AUUGCUUGGCAAGUAGUA----AACUGCUUGGCAAGCA ........---....-----...-.....-..((((((.(((((((..----..))))))).)))))) ( -19.40, z-score = -4.06, R) >consensus AGAUAUGAU_AUAUG_____AU__UAGAU_AUUGCUUGGCAAGUAGUA____AACUGCUUGGCAAGCA ................................((((((.((((((((......)))))))).)))))) (-16.71 = -16.93 + 0.23)

| Location | 19,172,574 – 19,172,637 |
|---|---|
| Length | 63 |
| Sequences | 3 |
| Columns | 68 |
| Reading direction | reverse |
| Mean pairwise identity | 64.77 |
| Shannon entropy | 0.46525 |
| G+C content | 0.37970 |
| Mean single sequence MFE | -16.10 |
| Consensus MFE | -11.45 |
| Energy contribution | -11.90 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.997193 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
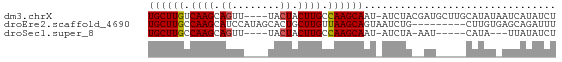
>dm3.chrX 19172574 63 - 22422827 UGCUUGUCAAGCAGUU----UACUACUUGCCAAGCAAU-AUCUACGAUGCUUGCAUAUAAUCAUAUCU ((((((.((((.((..----..)).)))).))))))..-.......(((....)))............ ( -13.50, z-score = -1.66, R) >droEre2.scaffold_4690 9430753 59 - 18748788 UGCUUGCCAAGCAUCCAUAGCACUGCUUGUUAAGCAGUAAUCUG---------CUUGUGAGCAGAUUU ((((((.((((((..........)))))).))))))..((((((---------((....)))))))). ( -21.90, z-score = -2.82, R) >droSec1.super_8 1467207 54 - 3762037 UGCUUGCCAAGCAGUU----UACUACUUGCCAAGCAAU-AUCUA-AAU-----CAUA---UUAUAUCU ((((((.((((.((..----..)).)))).))))))..-.....-...-----....---........ ( -12.90, z-score = -2.98, R) >consensus UGCUUGCCAAGCAGUU____UACUACUUGCCAAGCAAU_AUCUA__AU_____CAUAU_AUCAUAUCU ((((((.((((.(((......))).)))).))))))................................ (-11.45 = -11.90 + 0.45)
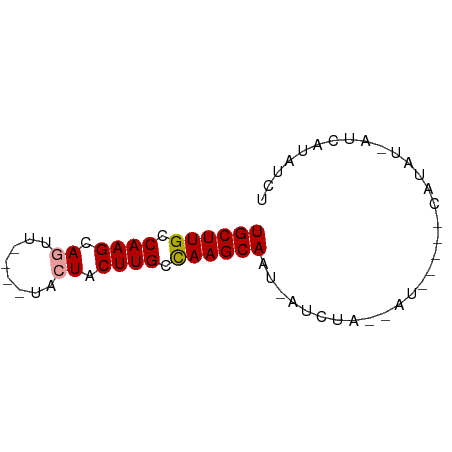

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:52 2011