| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,170,433 – 19,170,493 |
| Length | 60 |
| Max. P | 0.500000 |

| Location | 19,170,433 – 19,170,493 |
|---|---|
| Length | 60 |
| Sequences | 5 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 61.72 |
| Shannon entropy | 0.62248 |
| G+C content | 0.51805 |
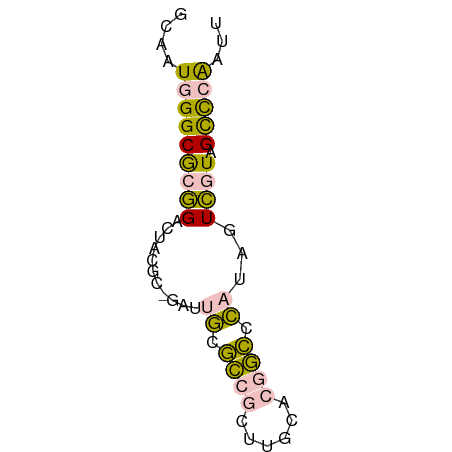
| Mean single sequence MFE | -17.30 |
| Consensus MFE | -9.00 |
| Energy contribution | -9.16 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.59 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
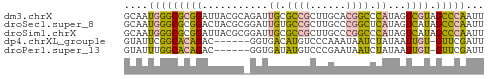
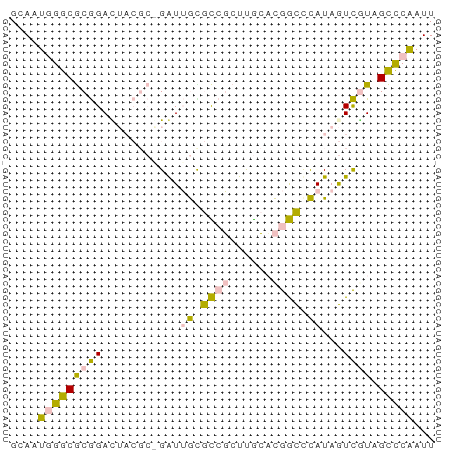
>dm3.chrX 19170433 60 - 22422827 GCAAUGGGCGCGGAUUACGCAGAUUGCGCCGCUUGCACGGCCCAUAGUCGUAGCCCAAUU ....(((((((((....(((.....)))))))....(((((.....))))).)))))... ( -23.50, z-score = -1.42, R) >droSec1.super_8 1464771 60 - 3762037 GCAAUGGGCGCGGACUACGCGGAUUGUGCCGCUUGCCCGGCUCAUAGUCAUAGCCCAAUU ....((((((((.....))).((((((((((......))))..))))))...)))))... ( -24.20, z-score = -1.61, R) >droSim1.chrX 14821823 60 - 17042790 GCAAUGGGCGCGGAUUACGCGGAUUGCGCCGCUUGCCCGGCCCAUAGUCAUAGCCCAAUU ....((((((((.....))).(((((.((((......))))...)))))...)))))... ( -22.70, z-score = -0.60, R) >dp4.chrXL_group1e 4116922 53 + 12523060 GUAUUCGGCACAGAC------GGUGACAUGUCCCAAAUAAUCUAUAAUUGU-GUUCGAUU ....(((((((((..------((.((....))))..(((...)))..))))-).)))).. ( -8.10, z-score = 0.28, R) >droPer1.super_13 163429 53 - 2293547 GUAUUUGGCACAGAC------GGUGAUAUGUCCCGAAUAAUCUAUAAUUGU-GUUCGAUU .(((((((....(((------(......)))))))))))......(((((.-...))))) ( -8.00, z-score = 0.41, R) >consensus GCAAUGGGCGCGGACUACGC_GAUUGCGCCGCUUGCACGGCCCAUAGUCGUAGCCCAAUU ....((((((((.....))).(((((.((((......))))...)))))...)))))... ( -9.00 = -9.16 + 0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:51 2011