| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,966,772 – 11,966,858 |
| Length | 86 |
| Max. P | 0.972208 |

| Location | 11,966,772 – 11,966,858 |
|---|---|
| Length | 86 |
| Sequences | 12 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 75.77 |
| Shannon entropy | 0.47075 |
| G+C content | 0.45523 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -9.58 |
| Energy contribution | -9.28 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
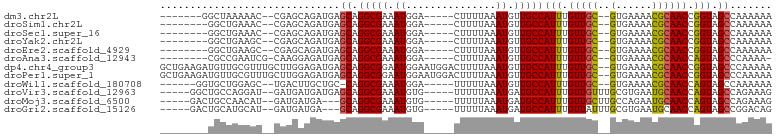
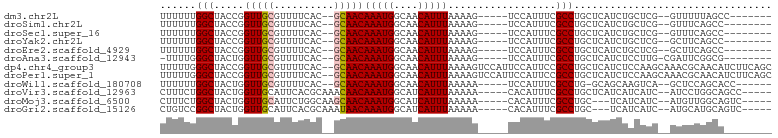
>dm3.chr2L 11966772 86 + 23011544 -------GGCUAAAAAC--CGAGCAGAUGAGCAGGCGAAAUGGA-----CUUUUAAAUGUUGCCAUUUGUUGC--GUGAAAACGCAACCGGUAGCCAAAAAA -------((((....((--((.((......)).(((((.((...-----.......)).)))))....(((((--((....)))))))))))))))...... ( -28.50, z-score = -3.27, R) >droSim1.chr2L 11773821 85 + 22036055 --------GGCUGAAAC--CGAGCAGAUGAGCAGGCGAAAUGGA-----CUUUUAAAUGUUGCCAUUUGUUGC--GUGAAAACGCAACCGGUAGCCAAAAAA --------((((...((--((.((......)).(((((.((...-----.......)).)))))....(((((--((....)))))))))))))))...... ( -28.90, z-score = -3.23, R) >droSec1.super_16 166134 85 + 1878335 --------GGCUGAAAC--CGAGCAGAUGAGCAGGCGAAAUGGA-----CUUUUAAAUGUUGCCAUUUGUUGC--GUGAAAACGCAACCGGUAGCCAAAAAA --------((((...((--((.((......)).(((((.((...-----.......)).)))))....(((((--((....)))))))))))))))...... ( -28.90, z-score = -3.23, R) >droYak2.chr2L 8388867 85 + 22324452 --------GGCUGAAGC--CGAGCAGAUGAGCAGGCGAAAUGGA-----CUUUUAAAUGUUGCCAUUUGUUGC--GUGAAAACGCAACCGGUAGCCAAAAAA --------(((((..((--(..((......)).))).(((((((-----(........))..))))))(((((--((....)))))))...)))))...... ( -29.60, z-score = -3.00, R) >droEre2.scaffold_4929 13197742 85 - 26641161 --------GGCUGAAGC--CGAGCAGAUGAGCAGGCGAAAUGGA-----CUUUUAAAUGUUGCCAUUUGUUGC--GUGAAAACGCAACCGGUAGCCAAAAAA --------(((((..((--(..((......)).))).(((((((-----(........))..))))))(((((--((....)))))))...)))))...... ( -29.60, z-score = -3.00, R) >droAna3.scaffold_12943 2762928 85 - 5039921 --------CGCCGAAUCG-CAAGGAGAUGAGCAGGCGAAAUGGA-----CUUUUAAAUGUUGCCAUUUGUUGC--GUGAAAACGCAACCAGUAGCCCAAAA- --------.((((..((.-...))...)).)).(((.(((((((-----(........))..))))))(((((--((....))))))).....))).....- ( -24.20, z-score = -1.66, R) >dp4.chr4_group3 4383925 100 - 11692001 GCUGAAGAUGUUGCGUUUGCUUGGAGAUGAGCAGGCGGAAUGGAAUGGACUUUUAAAUGUUGCCAUUUGUUGC--GUGAAAACGCAACCGGUAGCCCAAAAA ((((..(..(((.(((((((((......))))))))).))).((((((((........))..))))))(((((--((....))))))))..))))....... ( -33.70, z-score = -2.86, R) >droPer1.super_1 1529224 100 - 10282868 GCUGAAGAUGUUGCGUUUGCUUGGAGAUGAGCAGGCGGAAUGGAAUGGACUUUUAAAUGUUGCCAUUUGUUGC--GUGAAAACGCAACCGGUAGCCCAAAAA ((((..(..(((.(((((((((......))))))))).))).((((((((........))..))))))(((((--((....))))))))..))))....... ( -33.70, z-score = -2.86, R) >droWil1.scaffold_180708 2672726 86 + 12563649 ------GGUGCUGGAGC--UGACUUGCUGC-CAGGCGAAAUGGA-----UUUUUAAAUGUUGCCAUUUGUUGC--GUGAAAACGCAACCAGUAGCCAAAAAA ------(((.(((((((--......))).)-)))((.((((((.-----..(......)...))))))(((((--((....)))))))..)).)))...... ( -28.20, z-score = -2.14, R) >droVir3.scaffold_12963 6983646 90 - 20206255 -----GGCUGCCAGGAU--GAUGAUGAUGAGCAGGCGAAAUGUG-----UUUUUAAAUGAUGCCAUUUGUUGUUUGCGUGAAUGCAACCAGUAGCCAGAAAG -----((((((..((.(--(..........((((((((((((.(-----(.((.....)).))))))..)))))))).......)).)).))))))...... ( -22.73, z-score = -0.86, R) >droMoj3.scaffold_6500 15584421 87 - 32352404 -----GACUGCCAACAU--GAUGAUGA---GCAGGCGAAAUGUG-----UUUUUAAAUGAUGCCAUUUGUUGCUUGCCAGAAUGCAACCAGUAGCCAGAAAG -----.((((....(((--..((..((---((((.((((..(((-----((.......)))))..))))))))))..))..)))....)))).......... ( -18.10, z-score = -0.06, R) >droGri2.scaffold_15126 6635597 87 + 8399593 -----GACUGCAUGCAU--GAUGAUGA---GCAGGCGAAAUGUG-----UUUUUAAAUGAUGCCAUUUGUUAUUUGCGUGAAUGCAACCAGUAGCCGGACAG -----..(((((((((.--(((((..(---(..((((..((...-----.......))..)))).))..))))))))))).......((.......)).))) ( -17.30, z-score = 1.14, R) >consensus ________UGCUGAAAC__CGAGCAGAUGAGCAGGCGAAAUGGA_____CUUUUAAAUGUUGCCAUUUGUUGC__GUGAAAACGCAACCGGUAGCCAAAAAA .................................(((....((............(((((....)))))(((((..........)))))))...)))...... ( -9.58 = -9.28 + -0.30)
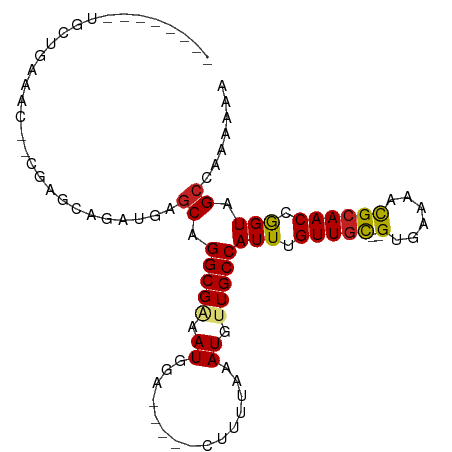
| Location | 11,966,772 – 11,966,858 |
|---|---|
| Length | 86 |
| Sequences | 12 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 75.77 |
| Shannon entropy | 0.47075 |
| G+C content | 0.45523 |
| Mean single sequence MFE | -22.43 |
| Consensus MFE | -12.55 |
| Energy contribution | -12.64 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.972208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
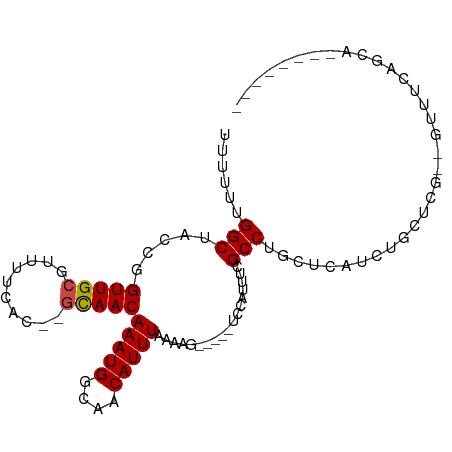
>dm3.chr2L 11966772 86 - 23011544 UUUUUUGGCUACCGGUUGCGUUUUCAC--GCAACAAAUGGCAACAUUUAAAAG-----UCCAUUUCGCCUGCUCAUCUGCUCG--GUUUUUAGCC------- ......(((((((((((((((....))--)))))(((((....))))).....-----............((......)).))--))....))))------- ( -23.90, z-score = -2.74, R) >droSim1.chr2L 11773821 85 - 22036055 UUUUUUGGCUACCGGUUGCGUUUUCAC--GCAACAAAUGGCAACAUUUAAAAG-----UCCAUUUCGCCUGCUCAUCUGCUCG--GUUUCAGCC-------- ......(((((((((((((((....))--)))))(((((....))))).....-----............((......)).))--))...))))-------- ( -24.30, z-score = -2.89, R) >droSec1.super_16 166134 85 - 1878335 UUUUUUGGCUACCGGUUGCGUUUUCAC--GCAACAAAUGGCAACAUUUAAAAG-----UCCAUUUCGCCUGCUCAUCUGCUCG--GUUUCAGCC-------- ......(((((((((((((((....))--)))))(((((....))))).....-----............((......)).))--))...))))-------- ( -24.30, z-score = -2.89, R) >droYak2.chr2L 8388867 85 - 22324452 UUUUUUGGCUACCGGUUGCGUUUUCAC--GCAACAAAUGGCAACAUUUAAAAG-----UCCAUUUCGCCUGCUCAUCUGCUCG--GCUUCAGCC-------- ......((((....(((((((....))--)))))(((((....))))).....-----........(((.((......))..)--))...))))-------- ( -25.00, z-score = -2.91, R) >droEre2.scaffold_4929 13197742 85 + 26641161 UUUUUUGGCUACCGGUUGCGUUUUCAC--GCAACAAAUGGCAACAUUUAAAAG-----UCCAUUUCGCCUGCUCAUCUGCUCG--GCUUCAGCC-------- ......((((....(((((((....))--)))))(((((....))))).....-----........(((.((......))..)--))...))))-------- ( -25.00, z-score = -2.91, R) >droAna3.scaffold_12943 2762928 85 + 5039921 -UUUUGGGCUACUGGUUGCGUUUUCAC--GCAACAAAUGGCAACAUUUAAAAG-----UCCAUUUCGCCUGCUCAUCUCCUUG-CGAUUCGGCG-------- -...((((((....(((((((....))--)))))(((((....)))))...))-----))))...(((((((..........)-))....))))-------- ( -27.30, z-score = -3.51, R) >dp4.chr4_group3 4383925 100 + 11692001 UUUUUGGGCUACCGGUUGCGUUUUCAC--GCAACAAAUGGCAACAUUUAAAAGUCCAUUCCAUUCCGCCUGCUCAUCUCCAAGCAAACGCAACAUCUUCAGC ....((((((....(((((((....))--)))))(((((....)))))...)))))).........((.((((........))))...))............ ( -26.10, z-score = -3.42, R) >droPer1.super_1 1529224 100 + 10282868 UUUUUGGGCUACCGGUUGCGUUUUCAC--GCAACAAAUGGCAACAUUUAAAAGUCCAUUCCAUUCCGCCUGCUCAUCUCCAAGCAAACGCAACAUCUUCAGC ....((((((....(((((((....))--)))))(((((....)))))...)))))).........((.((((........))))...))............ ( -26.10, z-score = -3.42, R) >droWil1.scaffold_180708 2672726 86 - 12563649 UUUUUUGGCUACUGGUUGCGUUUUCAC--GCAACAAAUGGCAACAUUUAAAAA-----UCCAUUUCGCCUG-GCAGCAAGUCA--GCUCCAGCACC------ .....(((......(((((((....))--)))))(((((....))))).....-----.)))....((.((-(.(((......--))))))))...------ ( -25.10, z-score = -2.61, R) >droVir3.scaffold_12963 6983646 90 + 20206255 CUUUCUGGCUACUGGUUGCAUUCACGCAAACAACAAAUGGCAUCAUUUAAAAA-----CACAUUUCGCCUGCUCAUCAUCAUC--AUCCUGGCAGCC----- ......((((.(..(((((......)))).........(((............-----........)))..............--...)..).))))----- ( -13.35, z-score = 0.31, R) >droMoj3.scaffold_6500 15584421 87 + 32352404 CUUUCUGGCUACUGGUUGCAUUCUGGCAAGCAACAAAUGGCAUCAUUUAAAAA-----CACAUUUCGCCUGC---UCAUCAUC--AUGUUGGCAGUC----- .......((((...(((((..........)))))...))))............-----........(((.((---........--..)).)))....----- ( -15.80, z-score = 0.58, R) >droGri2.scaffold_15126 6635597 87 - 8399593 CUGUCCGGCUACUGGUUGCAUUCACGCAAAUAACAAAUGGCAUCAUUUAAAAA-----CACAUUUCGCCUGC---UCAUCAUC--AUGCAUGCAGUC----- ((((..(((......((((......)))).....(((((....))))).....-----........)))(((---........--..))).))))..----- ( -12.90, z-score = 0.98, R) >consensus UUUUUUGGCUACCGGUUGCGUUUUCAC__GCAACAAAUGGCAACAUUUAAAAG_____UCCAUUUCGCCUGCUCAUCUGCUCG__GUUUCAGCA________ ......(((.....(((((..........)))))(((((....)))))..................)))................................. (-12.55 = -12.64 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:03 2011