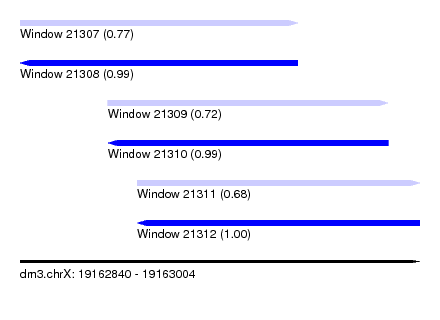
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,162,840 – 19,163,004 |
| Length | 164 |
| Max. P | 0.997703 |

| Location | 19,162,840 – 19,162,954 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 90.22 |
| Shannon entropy | 0.16843 |
| G+C content | 0.49915 |
| Mean single sequence MFE | -41.96 |
| Consensus MFE | -31.94 |
| Energy contribution | -32.70 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.773085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
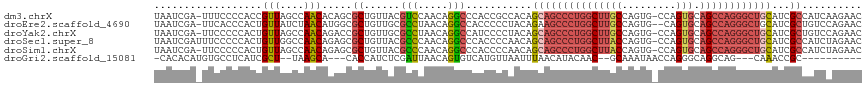
>dm3.chrX 19162840 114 + 22422827 AAAUACACAGCGAGUAUUUUCCUUU----UUUUUAUUUGUUCUGCCACAGAUGUUCUUGAUGGCGAUGCAGCCCUGGCUGCACUGGCACUGGCAAGCCAGGGCUGCUGUGGCGGUGGG .....(((((((((((.........----....)))))))).(((((((..((((......))))..((((((((((((((.(.......))).)))))))))))))))))))))).. ( -45.92, z-score = -2.58, R) >droEre2.scaffold_4690 9421725 117 + 18748788 AAAUACACAGCCACUAUUUUCUUUUCUUCUUUUUAUAUGUUCUGCCACAGAUGUUCUGGACAGCGAUGCAGCCCUGGCUGCACUG-CACUGGCAAGCCAGGGCUUCUGUAGGGGGUGG .....(((..((.((((....................(((..(((..(((.....)))....)))..)))(((((((((((.(..-....))).)))))))))....)))))).))). ( -37.30, z-score = -0.89, R) >droYak2.chrX 9607911 112 - 21770863 AAAUACACAGCCACUAUAUUUUU------UUUUUAUUUGUUCUGCCACAGAUGUUCUGGACAGCGAUGCAGCCCUGGCUGCACUGGCACUGGCAAGCCAGGGCUGCUGUAGGGGAUGG .......((.((.(((((.....------.........((((((.(.(((.....)))).))).)))((((((((((((((.(.......))).))))))))))))))))).)).)). ( -41.00, z-score = -1.99, R) >droSec1.super_8 1457163 114 + 3762037 AAAUACACAGCGACUAUUUUUCUUU----UUUUUAUUUGUUCUGCCACAGAUGUUCUAGAUGGCGAUGCAGCCCUGGCUGCACUGGCACUGGUAAGCCAGGGCUGCUGUUGGGGUGGG .....(((..((((...........----.............((((((((.....)).).)))))..((((((((((((..(((......))).)))))))))))).))))..))).. ( -42.80, z-score = -2.66, R) >droSim1.chrX 14814299 114 + 17042790 AAAUACACAGCGACUAUUUUUCUUU----UUUUUAUUUGUUCUGCCACAGAUGUUCUAGAUGGCGAUGCAGCCCUGGCUGCACUGGCACUGGUAAGCCAGGGCUGCUGUUGGGGUGGG .....(((..((((...........----.............((((((((.....)).).)))))..((((((((((((..(((......))).)))))))))))).))))..))).. ( -42.80, z-score = -2.66, R) >consensus AAAUACACAGCGACUAUUUUUCUUU____UUUUUAUUUGUUCUGCCACAGAUGUUCUAGAUGGCGAUGCAGCCCUGGCUGCACUGGCACUGGCAAGCCAGGGCUGCUGUAGGGGUGGG ......(((((...........................((..((((((((.....)).).)))))..))((((((((((((.(.......))).)))))))))))))))......... (-31.94 = -32.70 + 0.76)

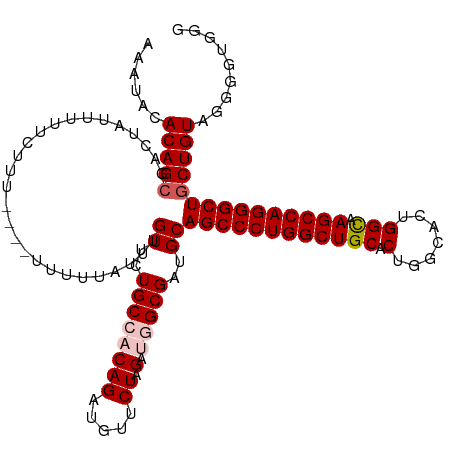
| Location | 19,162,840 – 19,162,954 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 90.22 |
| Shannon entropy | 0.16843 |
| G+C content | 0.49915 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -33.70 |
| Energy contribution | -33.54 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.994814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19162840 114 - 22422827 CCCACCGCCACAGCAGCCCUGGCUUGCCAGUGCCAGUGCAGCCAGGGCUGCAUCGCCAUCAAGAACAUCUGUGGCAGAACAAAUAAAAA----AAAGGAAAAUACUCGCUGUGUAUUU ......((((((((((((((((((.(((.......).)))))))))))))).((........)).....))))))..............----......((((((.......)))))) ( -42.10, z-score = -3.28, R) >droEre2.scaffold_4690 9421725 117 - 18748788 CCACCCCCUACAGAAGCCCUGGCUUGCCAGUG-CAGUGCAGCCAGGGCUGCAUCGCUGUCCAGAACAUCUGUGGCAGAACAUAUAAAAAGAAGAAAAGAAAAUAGUGGCUGUGUAUUU .(((...((((.(.((((((((((.(((....-..).)))))))))))).)....((((((((.....))).)))))...........................))))..)))..... ( -38.40, z-score = -2.16, R) >droYak2.chrX 9607911 112 + 21770863 CCAUCCCCUACAGCAGCCCUGGCUUGCCAGUGCCAGUGCAGCCAGGGCUGCAUCGCUGUCCAGAACAUCUGUGGCAGAACAAAUAAAAA------AAAAAUAUAGUGGCUGUGUAUUU .(((...((((.((((((((((((.(((.......).))))))))))))))....((((((((.....))).)))))............------.........))))..)))..... ( -43.00, z-score = -3.52, R) >droSec1.super_8 1457163 114 - 3762037 CCCACCCCAACAGCAGCCCUGGCUUACCAGUGCCAGUGCAGCCAGGGCUGCAUCGCCAUCUAGAACAUCUGUGGCAGAACAAAUAAAAA----AAAGAAAAAUAGUCGCUGUGUAUUU ..(((..(.((.(((((((((((((((........))).))))))))))))...(((((..((.....)))))))..............----...........)).)..)))..... ( -38.00, z-score = -3.63, R) >droSim1.chrX 14814299 114 - 17042790 CCCACCCCAACAGCAGCCCUGGCUUACCAGUGCCAGUGCAGCCAGGGCUGCAUCGCCAUCUAGAACAUCUGUGGCAGAACAAAUAAAAA----AAAGAAAAAUAGUCGCUGUGUAUUU ..(((..(.((.(((((((((((((((........))).))))))))))))...(((((..((.....)))))))..............----...........)).)..)))..... ( -38.00, z-score = -3.63, R) >consensus CCCACCCCAACAGCAGCCCUGGCUUGCCAGUGCCAGUGCAGCCAGGGCUGCAUCGCCAUCCAGAACAUCUGUGGCAGAACAAAUAAAAA____AAAGAAAAAUAGUCGCUGUGUAUUU ........(((((((((((((((((((........))).)))))))))).....((((..(((.....)))))))................................))))))..... (-33.70 = -33.54 + -0.16)

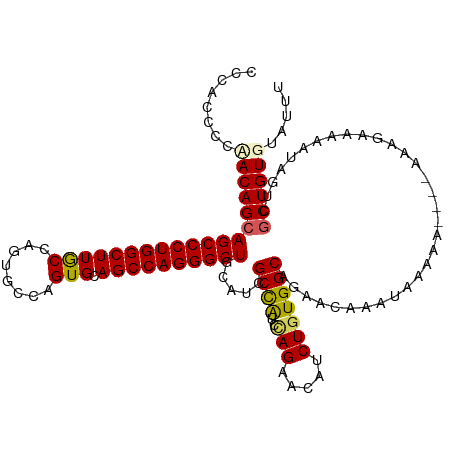
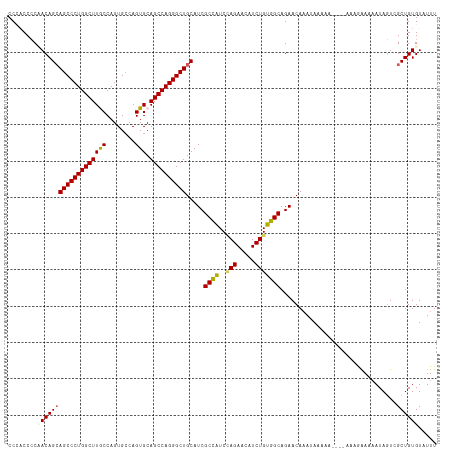
| Location | 19,162,876 – 19,162,991 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 73.97 |
| Shannon entropy | 0.50602 |
| G+C content | 0.57840 |
| Mean single sequence MFE | -46.93 |
| Consensus MFE | -29.48 |
| Energy contribution | -30.77 |
| Covariance contribution | 1.29 |
| Combinations/Pair | 1.44 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.724112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19162876 115 + 22422827 UCUGCCACAGAUGUUCUUGAUGGCGAUGCAGCCCUGGCUGCACUGGCACUGGCAAGCCAGGGCUGCUGUGGCGGUGGGCCUGUUGGACGUAACAGCGCUGUGUUGGCUAACGGUG .((((((((..((((......))))..((((((((((((((.(.......))).))))))))))))))))))))...(((.(((((.(.(((((......)))))))))))))). ( -51.90, z-score = -1.09, R) >droEre2.scaffold_4690 9421765 114 + 18748788 UCUGCCACAGAUGUUCUGGACAGCGAUGCAGCCCUGGCUGCACUG-CACUGGCAAGCCAGGGCUUCUGUAGGGGGUGGCCUGUUAGGCGCAACAGCGCCAUGUUAGAUAACAGUG .((.(((((((.(((((((.(((.(.((((((....))))))...-).))).....))))))).))))).)).))....(((((((((((....)))))........)))))).. ( -48.70, z-score = -0.98, R) >droYak2.chrX 9607945 115 - 21770863 UCUGCCACAGAUGUUCUGGACAGCGAUGCAGCCCUGGCUGCACUGGCACUGGCAAGCCAGGGCUGCUGUAGGGGAUGGCCUGUUAGGCGCAACAGCGGUCUGUUGGCUAACAGUG ...(((((((.....)))..(.(((..((((((((((((((.(.......))).))))))))))))))).)....))))(((((((.(.((((((....)))))))))))))).. ( -52.00, z-score = -0.99, R) >droSec1.super_8 1457199 115 + 3762037 UCUGCCACAGAUGUUCUAGAUGGCGAUGCAGCCCUGGCUGCACUGGCACUGGUAAGCCAGGGCUGCUGUUGGGGUGGGCCUGUUGGGCGUAACAGCGCUCUGUUGGCCAACAGUG ..((((((((.....)).).)))))..((((((((((((..(((......))).)))))))))))).......((.((((....((((((....))))))....)))).)).... ( -53.90, z-score = -1.42, R) >droSim1.chrX 14814335 115 + 17042790 UCUGCCACAGAUGUUCUAGAUGGCGAUGCAGCCCUGGCUGCACUGGCACUGGUAAGCCAGGGCUGCUGUUGGGGUGGGCCUGUUGGGCGUAACAGCGCUCUGUUGGCUAACAGUG ..((((((((.....)).).)))))..((((((((((((..(((......))).)))))))))))).......((.((((....((((((....))))))....)))).)).... ( -52.10, z-score = -1.26, R) >droGri2.scaffold_15081 1076400 96 + 4274704 ---GCUGUUGCUGUUGCGGUUUGCUGC-CUGCCCUGGUU--AUUUGCGUUG-UAUGUUAAA-------UUAACAUGACACUGUUAAUCGAGAUGGUGUGCU--UAAGCGAUG--- ---...((((((...((.....((.((-(......))).--....))....-(((((((..-------.)))))))((((((((......)))))))))).--..)))))).--- ( -23.00, z-score = -0.12, R) >consensus UCUGCCACAGAUGUUCUGGAUGGCGAUGCAGCCCUGGCUGCACUGGCACUGGCAAGCCAGGGCUGCUGUUGGGGUGGGCCUGUUAGGCGUAACAGCGCUCUGUUGGCUAACAGUG ...(((......(((......)))...((((((((((((((.(.......)))).)))))))))))..........)))(((((((...((((((....)))))).))))))).. (-29.48 = -30.77 + 1.29)
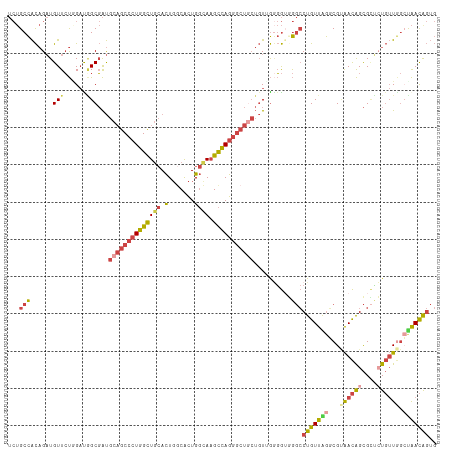
| Location | 19,162,876 – 19,162,991 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 73.97 |
| Shannon entropy | 0.50602 |
| G+C content | 0.57840 |
| Mean single sequence MFE | -43.02 |
| Consensus MFE | -28.08 |
| Energy contribution | -29.47 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.994845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
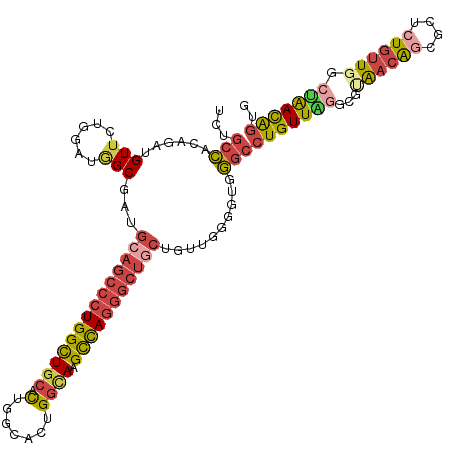
>dm3.chrX 19162876 115 - 22422827 CACCGUUAGCCAACACAGCGCUGUUACGUCCAACAGGCCCACCGCCACAGCAGCCCUGGCUUGCCAGUGCCAGUGCAGCCAGGGCUGCAUCGCCAUCAAGAACAUCUGUGGCAGA .................((.(((((......))))))).....((((((((((((((((((.(((.......).)))))))))))))).((........)).....))))))... ( -45.90, z-score = -2.23, R) >droEre2.scaffold_4690 9421765 114 - 18748788 CACUGUUAUCUAACAUGGCGCUGUUGCGCCUAACAGGCCACCCCCUACAGAAGCCCUGGCUUGCCAGUG-CAGUGCAGCCAGGGCUGCAUCGCUGUCCAGAACAUCUGUGGCAGA ..((((((........(((((....)))))))))))(((((.....((((.((((((((((.(((....-..).))))))))))))......))))..((.....)))))))... ( -47.50, z-score = -1.78, R) >droYak2.chrX 9607945 115 + 21770863 CACUGUUAGCCAACAGACCGCUGUUGCGCCUAACAGGCCAUCCCCUACAGCAGCCCUGGCUUGCCAGUGCCAGUGCAGCCAGGGCUGCAUCGCUGUCCAGAACAUCUGUGGCAGA ..((((((((((((((....)))))).).))))))).............((((((((((((.(((.......).))))))))))))))....((((((((.....))).))))). ( -55.70, z-score = -4.04, R) >droSec1.super_8 1457199 115 - 3762037 CACUGUUGGCCAACAGAGCGCUGUUACGCCCAACAGGCCCACCCCAACAGCAGCCCUGGCUUACCAGUGCCAGUGCAGCCAGGGCUGCAUCGCCAUCUAGAACAUCUGUGGCAGA ..((((((((.(((((....)))))..).))))))).............(((((((((((((((........))).))))))))))))...(((((..((.....)))))))... ( -49.70, z-score = -2.92, R) >droSim1.chrX 14814335 115 - 17042790 CACUGUUAGCCAACAGAGCGCUGUUACGCCCAACAGGCCCACCCCAACAGCAGCCCUGGCUUACCAGUGCCAGUGCAGCCAGGGCUGCAUCGCCAUCUAGAACAUCUGUGGCAGA ........((((.((((((.(((((......)))))))...........(((((((((((((((........))).))))))))))))................))))))))... ( -46.40, z-score = -2.76, R) >droGri2.scaffold_15081 1076400 96 - 4274704 ---CAUCGCUUA--AGCACACCAUCUCGAUUAACAGUGUCAUGUUAA-------UUUAACAUA-CAACGCAAAU--AACCAGGGCAG-GCAGCAAACCGCAACAGCAACAGC--- ---....(((..--.((...((......(((.....(((.((((((.-------..)))))))-)).....)))--......))...-)).((.....))...)))......--- ( -12.90, z-score = 0.63, R) >consensus CACUGUUAGCCAACAGAGCGCUGUUACGCCCAACAGGCCCACCCCAACAGCAGCCCUGGCUUACCAGUGCCAGUGCAGCCAGGGCUGCAUCGCCAUCCAGAACAUCUGUGGCAGA ........((((.((((...(((....(((.....)))...........(((((((((((((((........))).)))))))))))).........)))....))))))))... (-28.08 = -29.47 + 1.38)

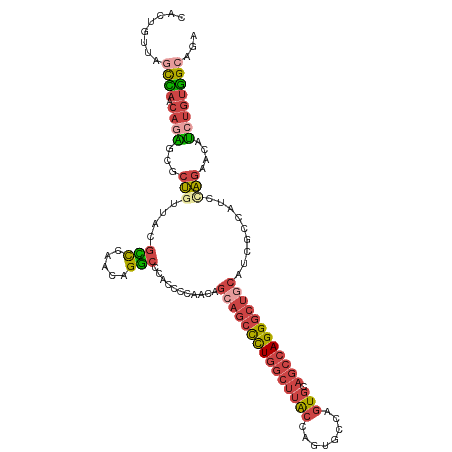
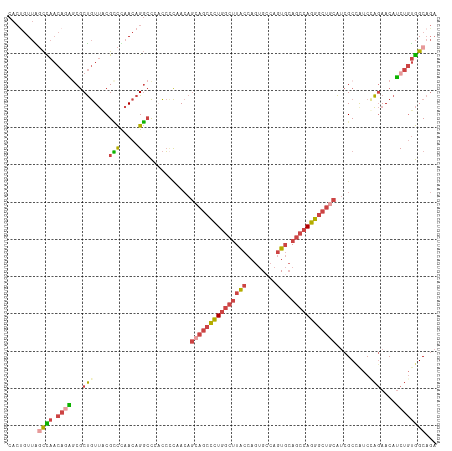
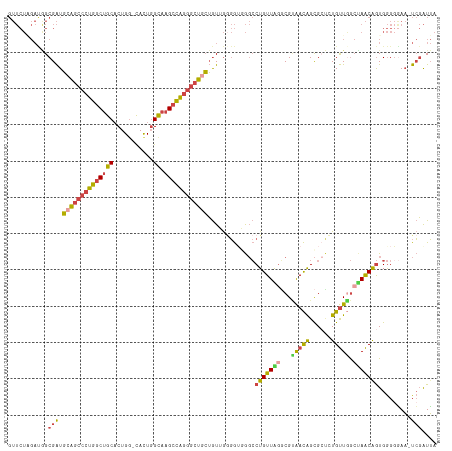
| Location | 19,162,888 – 19,163,004 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 72.69 |
| Shannon entropy | 0.53224 |
| G+C content | 0.57126 |
| Mean single sequence MFE | -44.32 |
| Consensus MFE | -29.63 |
| Energy contribution | -29.75 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.67 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.679446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19162888 116 + 22422827 GUUCUUGAUGGCGAUGCAGCCCUGGCUGCACUGG-CACUGGCAAGCCAGGGCUGCUGUGGCGGUGGGCCUGUUGGACGUAACAGCGCUGUGUUGGCUAACGGUGGGGAAA-UCGAUUA .(((((.((.((.((((((((((((((((.(...-....))).)))))))))))).)).)).((.((((...(((.(((....))))))....)))).)).)).))))).-....... ( -47.40, z-score = -0.62, R) >droEre2.scaffold_4690 9421777 115 + 18748788 GUUCUGGACAGCGAUGCAGCCCUGGCUGCACUG--CACUGGCAAGCCAGGGCUUCUGUAGGGGGUGGCCUGUUAGGCGCAACAGCGCCAUGUUAGAUAACAGUGGGUGAA-UCGAUUA (..((..((((......((((((((((((.(..--....))).)))))))))).))))..))..).(((..((.(((((....)))))..(((....)))))..)))...-....... ( -44.20, z-score = -0.24, R) >droYak2.chrX 9607957 116 - 21770863 GUUCUGGACAGCGAUGCAGCCCUGGCUGCACUGG-CACUGGCAAGCCAGGGCUGCUGUAGGGGAUGGCCUGUUAGGCGCAACAGCGGUCUGUUGGCUAACAGUGGGGGAA-UCGAUUA (((((...(((...((((((....))))))))).-(((((...(((((((((((((((......(((((.....))).))))))))))))..)))))..)))))..))))-)...... ( -48.70, z-score = -0.82, R) >droSec1.super_8 1457211 117 + 3762037 GUUCUAGAUGGCGAUGCAGCCCUGGCUGCACUGG-CACUGGUAAGCCAGGGCUGCUGUUGGGGUGGGCCUGUUGGGCGUAACAGCGCUCUGUUGGCCAACAGUGGGGGAAAUCGAUUA .((((...(.((...((((((((((((..(((..-....))).)))))))))))).......((.((((....((((((....))))))....)))).)).)).).))))........ ( -52.20, z-score = -1.63, R) >droSim1.chrX 14814347 116 + 17042790 GUUCUAGAUGGCGAUGCAGCCCUGGCUGCACUGG-CACUGGUAAGCCAGGGCUGCUGUUGGGGUGGGCCUGUUGGGCGUAACAGCGCUCUGUUGGCUAACAGUGGGGGAA-UCGAUUA (((((...(.((...((((((((((((..(((..-....))).)))))))))))).......((.((((....((((((....))))))....)))).)).)).).))))-)...... ( -50.90, z-score = -1.61, R) >droGri2.scaffold_15081 1076412 97 + 4274704 ----------GCGGUUUG---CUGCCUGCCCUGGUUAUUUGC--GUUGUAUGUUAAAUUAACAUGACACUGUUAAUCGAGAUGGUG---UGCUUA--AGCGAUGAGGCACAUGUGUG- ----------((((....---)))).((((((.(((....((--....(((((((...)))))))((((((((......)))))))---)))...--))).).).))))........- ( -22.50, z-score = 0.89, R) >consensus GUUCUAGAUGGCGAUGCAGCCCUGGCUGCACUGG_CACUGGCAAGCCAGGGCUGCUGUUGGGGUGGGCCUGUUAGGCGUAACAGCGCUCUGUUGGCUAACAGUGGGGGAA_UCGAUUA ...........(((.((((((((((((((...(....)..)).)))))))))))).............(((((((...(((((......))))).))))))).........))).... (-29.63 = -29.75 + 0.12)
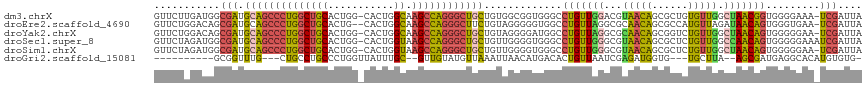
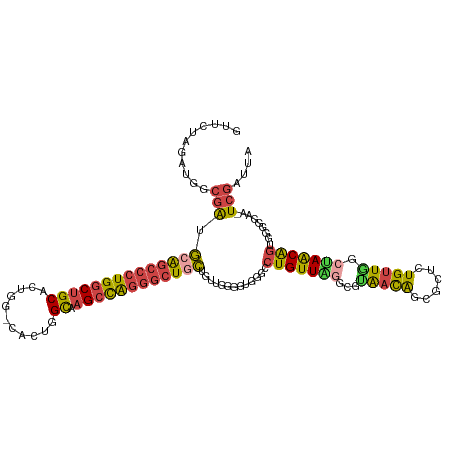
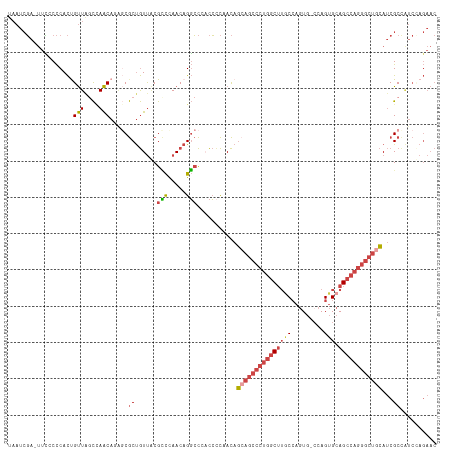
| Location | 19,162,888 – 19,163,004 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 72.69 |
| Shannon entropy | 0.53224 |
| G+C content | 0.57126 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -21.66 |
| Energy contribution | -22.92 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.997703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
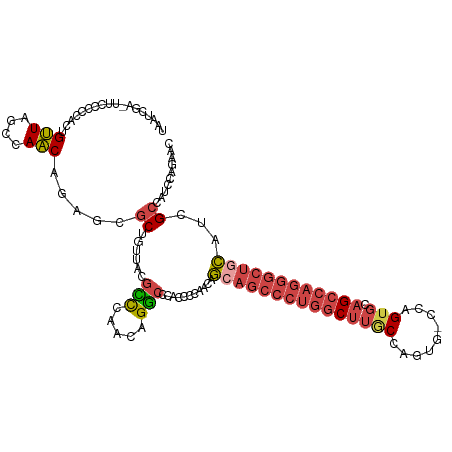
>dm3.chrX 19162888 116 - 22422827 UAAUCGA-UUUCCCCACCGUUAGCCAACACAGCGCUGUUACGUCCAACAGGCCCACCGCCACAGCAGCCCUGGCUUGCCAGUG-CCAGUGCAGCCAGGGCUGCAUCGCCAUCAAGAAC ....(((-(........((((((((......).))))..))).......(((.....)))...((((((((((((.(((....-...).))))))))))))))))))........... ( -38.30, z-score = -2.42, R) >droEre2.scaffold_4690 9421777 115 - 18748788 UAAUCGA-UUCACCCACUGUUAUCUAACAUGGCGCUGUUGCGCCUAACAGGCCACCCCCUACAGAAGCCCUGGCUUGCCAGUG--CAGUGCAGCCAGGGCUGCAUCGCUGUCCAGAAC .......-(((.....((((((........(((((....)))))))))))..........((((.((((((((((.(((....--..).))))))))))))......))))...))). ( -40.40, z-score = -1.72, R) >droYak2.chrX 9607957 116 + 21770863 UAAUCGA-UUCCCCCACUGUUAGCCAACAGACCGCUGUUGCGCCUAACAGGCCAUCCCCUACAGCAGCCCUGGCUUGCCAGUG-CCAGUGCAGCCAGGGCUGCAUCGCUGUCCAGAAC ....(((-(.......((((((((((((((....)))))).).))))))).............((((((((((((.(((....-...).))))))))))))))))))........... ( -47.70, z-score = -4.26, R) >droSec1.super_8 1457211 117 - 3762037 UAAUCGAUUUCCCCCACUGUUGGCCAACAGAGCGCUGUUACGCCCAACAGGCCCACCCCAACAGCAGCCCUGGCUUACCAGUG-CCAGUGCAGCCAGGGCUGCAUCGCCAUCUAGAAC ....((((........((((((((.(((((....)))))..).))))))).............(((((((((((((((.....-...))).))))))))))))))))........... ( -44.40, z-score = -3.85, R) >droSim1.chrX 14814347 116 - 17042790 UAAUCGA-UUCCCCCACUGUUAGCCAACAGAGCGCUGUUACGCCCAACAGGCCCACCCCAACAGCAGCCCUGGCUUACCAGUG-CCAGUGCAGCCAGGGCUGCAUCGCCAUCUAGAAC ....(((-(.......(((((.((.(((((....)))))..))..))))).............(((((((((((((((.....-...))).))))))))))))))))........... ( -40.30, z-score = -3.51, R) >droGri2.scaffold_15081 1076412 97 - 4274704 -CACACAUGUGCCUCAUCGCU--UAAGCA---CACCAUCUCGAUUAACAGUGUCAUGUUAAUUUAACAUACAAC--GCAAAUAACCAGGGCAGGCAG---CAAACCGC---------- -......(((((((..((((.--...)).---.........((((((((......))))))))...........--...........))..))))).---))......---------- ( -15.20, z-score = 0.47, R) >consensus UAAUCGA_UUCCCCCACUGUUAGCCAACAGAGCGCUGUUACGCCCAACAGGCCCACCCCAACAGCAGCCCUGGCUUGCCAGUG_CCAGUGCAGCCAGGGCUGCAUCGCCAUCCAGAAC ..................(((....))).....((......(((.....)))...........(((((((((((((((.........))).))))))))))))...)).......... (-21.66 = -22.92 + 1.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:50 2011