| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,160,313 – 19,160,419 |
| Length | 106 |
| Max. P | 0.847783 |

| Location | 19,160,313 – 19,160,419 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 59.77 |
| Shannon entropy | 0.71336 |
| G+C content | 0.54856 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -15.58 |
| Energy contribution | -14.62 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.41 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
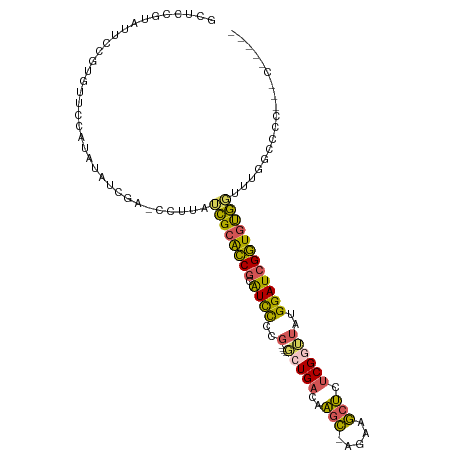
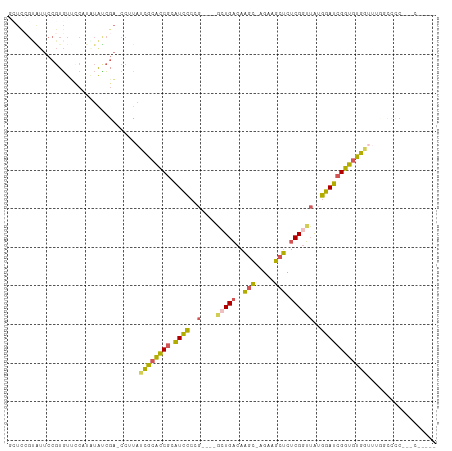
>dm3.chrX 19160313 106 - 22422827 GCUCCGUAUUCCGUGUUCCGUAUAUCGA-CCUUAUCGCACCGCAUCCCUGACAAGCUGACAAGC-AGAAGCUCUCGGUUAUGGAUCGGUGUGUUCUUGCCCCACCCUC--- ............(((...((.....)).-......(((((((.((((.((((.((((.......-...))))....)))).))))))))))).........)))....--- ( -24.40, z-score = -0.25, R) >droSec1.super_8 1454611 101 - 3762037 GCUCCGUAUUCCGUGUUCCGUGUAUCGACCCUUAUCGCACCGCAUCCCCG----GCUGACAAGC-AGAAGCUCUCGGUUAUGGAUCGGUGUGGCUUUCCCCCCCAC----- ((((.((((..((.....)).)))).)).......(((((((.((((..(----(((((..(((-....))).))))))..)))))))))))))............----- ( -27.20, z-score = -0.46, R) >droYak2.chrX 9605250 96 + 21770863 CGUUGGCGUUCCGUGUUCCAUGUGCCGA-CCUUAUCGCACCGCAUCCCCG----CCUGACAAGC-AGAAGCUCUCGAUUAUGGAUCGGUGUGGUUGGUUUUC--------- .(((((((...((((...))))))))))-)((.(((((((((.((((...----..(((..(((-....))).))).....))))))))))))).)).....--------- ( -31.40, z-score = -1.24, R) >droEre2.scaffold_4690 9419179 90 - 18748788 GUUUCGUGUUCCGUGUGGCCU-UAUCGA-CCUUAUCGCACCGCAUCCCCG----GCUGACAACC-AGAAGCUCUCGGUUAUGGAUCGGUGUGGUUUC-------------- ...(((....((....))...-...)))-....(((((((((.((((..(----(((((.....-........))))))..)))))))))))))...-------------- ( -23.22, z-score = 0.55, R) >droAna3.scaffold_12903 518929 91 - 802071 -------------UAUCUGACAAAUUGGACGAUAGUGGGUCAUGUUU-------UGUGUCAGGUGGGAAACCCUCGGGAGUGGAUCGAUACACGAUGGCUCCUAUCUACUC -------------.((((((((.....(((........)))......-------..))))))))((....))...((((((.(.(((.....)))).))))))........ ( -27.12, z-score = -0.67, R) >consensus GCUCCGUAUUCCGUGUUCCAUAUAUCGA_CCUUAUCGCACCGCAUCCCCG____GCUGACAAGC_AGAAGCUCUCGGUUAUGGAUCGGUGUGGUUUGGCCCC___C_____ ..................................((((((((.((((.......(((((..(((.....))).)))))...)))))))))))).................. (-15.58 = -14.62 + -0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:45 2011