| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,160,029 – 19,160,230 |
| Length | 201 |
| Max. P | 0.997205 |

| Location | 19,160,029 – 19,160,121 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.23 |
| Shannon entropy | 0.28990 |
| G+C content | 0.60191 |
| Mean single sequence MFE | -43.00 |
| Consensus MFE | -24.70 |
| Energy contribution | -25.03 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19160029 92 + 22422827 --------GCUGCGUCUGCGG----CAUUCCCAUUCUGAUUCCGCCUGGCGGUUCCUACAAGGCCGCCACCAGAGGUUCCAUAACGGGAAUGCAACGGAGCAGC------ --------(((((.((((..(----(((((((.((.((...((..((((.(((.((.....))..))).)))).))...)).)).))))))))..)))))))))------ ( -44.10, z-score = -4.18, R) >droSec1.super_8 1454332 97 + 3762037 AGCGCUCUGCUGCGGCUGCGG----CAUUCCCAUUCUGAUUCCGCCAGGCGGUUCCUACAAGGCCGCCACCAGA---UCCAUAACGGGAAUGUAUCGGAGCAGC------ ........(((((..(((..(----(((((((.((.((..((.(...(((((((.......)))))))..).))---..)).)).))))))))..))).)))))------ ( -36.70, z-score = -1.02, R) >droEre2.scaffold_4690 9418889 107 + 18748788 CGCGCUCUUCUGCGGAUGGCAUUCCCAUUCCCAUUCUGAUUCUGGCUGGCGGUUCCUACAAGGCCGCCACCAGA---UCCAAAACGGGAAUGCAACGGGGCAGCGAUGUC ((((((((..((((((......))).((((((.((.((..(((((.((((((((.......)))))))))))))---..)).)).)))))))))..))))).)))..... ( -48.20, z-score = -3.69, R) >consensus _GCGCUCUGCUGCGGCUGCGG____CAUUCCCAUUCUGAUUCCGCCUGGCGGUUCCUACAAGGCCGCCACCAGA___UCCAUAACGGGAAUGCAACGGAGCAGC______ ........(((((...((.......(((((((...............(((((((.......)))))))....(......).....)))))))...))..)))))...... (-24.70 = -25.03 + 0.33)


| Location | 19,160,029 – 19,160,121 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.23 |
| Shannon entropy | 0.28990 |
| G+C content | 0.60191 |
| Mean single sequence MFE | -46.77 |
| Consensus MFE | -32.40 |
| Energy contribution | -32.51 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.06 |
| SVM RNA-class probability | 0.997205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19160029 92 - 22422827 ------GCUGCUCCGUUGCAUUCCCGUUAUGGAACCUCUGGUGGCGGCCUUGUAGGAACCGCCAGGCGGAAUCAGAAUGGGAAUG----CCGCAGACGCAGC-------- ------(((((((.((.(((((((((((.(((..((((((((((...((.....))..)))))))).))..))).))))))))))----).)).)).)))))-------- ( -48.70, z-score = -4.76, R) >droSec1.super_8 1454332 97 - 3762037 ------GCUGCUCCGAUACAUUCCCGUUAUGGA---UCUGGUGGCGGCCUUGUAGGAACCGCCUGGCGGAAUCAGAAUGGGAAUG----CCGCAGCCGCAGCAGAGCGCU ------((.(((.((...((((((((((.(((.---((((.((((((((.....))..))))).).)))).))).))))))))))----.)).))).))(((.....))) ( -40.50, z-score = -1.35, R) >droEre2.scaffold_4690 9418889 107 - 18748788 GACAUCGCUGCCCCGUUGCAUUCCCGUUUUGGA---UCUGGUGGCGGCCUUGUAGGAACCGCCAGCCAGAAUCAGAAUGGGAAUGGGAAUGCCAUCCGCAGAAGAGCGCG .....(((.((.(..(((((((((((((((((.---(((((((((((((.....))..)))))).))))).))))))))))))).(((......)))))))..).))))) ( -51.10, z-score = -4.24, R) >consensus ______GCUGCUCCGUUGCAUUCCCGUUAUGGA___UCUGGUGGCGGCCUUGUAGGAACCGCCAGGCGGAAUCAGAAUGGGAAUG____CCGCAGCCGCAGCAGAGCGC_ ......(((((.......((((((((((...((...((((..(((((((.....))..)))))...)))).))..))))))))))............)))))........ (-32.40 = -32.51 + 0.11)


| Location | 19,160,047 – 19,160,155 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 68.34 |
| Shannon entropy | 0.49832 |
| G+C content | 0.56648 |
| Mean single sequence MFE | -37.37 |
| Consensus MFE | -20.10 |
| Energy contribution | -22.98 |
| Covariance contribution | 2.87 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.851204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19160047 108 - 22422827 GGUGUCAAGUUGGGUUCGAUAACCCAGCGC------CACCGCUGCUCCGUUGCAUUCCCGUUAUGGAACCUCUGGUGGCGGCCUUGUAGGAACCGCCAGGCGGAAUCAGAAUGG ((((....((((((((....))))))))..------))))..(((......)))...(((((.(((..((((((((((...((.....))..)))))))).))..))).))))) ( -42.10, z-score = -1.28, R) >droSec1.super_8 1454358 105 - 3762037 GGUGUCAAGUUGGGUUCGAUAACCCAGCGC------CACCGCUGCUCCGAUACAUUCCCGUUAUGGA---UCUGGUGGCGGCCUUGUAGGAACCGCCUGGCGGAAUCAGAAUGG .(((((..((((((((....))))))))((------....))......)))))....(((((.(((.---((((.((((((((.....))..))))).).)))).))).))))) ( -38.50, z-score = -0.79, R) >droEre2.scaffold_4690 9418919 111 - 18748788 GGUGUCAAGUAGGGGUCGAUAACCCAUCGCCAUCGACAUCGCUGCCCCGUUGCAUUCCCGUUUUGGA---UCUGGUGGCGGCCUUGUAGGAACCGCCAGCCAGAAUCAGAAUGG ((((((..((..((((.....))))...))....))))))((.((...)).))....(((((((((.---(((((((((((((.....))..)))))).))))).))))))))) ( -43.90, z-score = -2.19, R) >droAna3.scaffold_12903 518696 95 - 802071 GGUGUCAAGAGUGGGUCGAUAACCCAUUC-------AAUCCAAGC---------UUCCCAUUCCAAG---UUCCUCCGCCCAGUCGAGAUAAACGCCGAGGAAUACGAGAAGUG ........((((((((.....))))))))-------.......((---------(((....((...(---((((((.((...............)).)))))))..))))))). ( -24.96, z-score = -1.94, R) >consensus GGUGUCAAGUUGGGGUCGAUAACCCAGCGC______CACCGCUGCUCCGUUGCAUUCCCGUUAUGGA___UCUGGUGGCGGCCUUGUAGGAACCGCCAGGCAGAAUCAGAAUGG ((((....((((((........))))))........)))).................(((((((((....(((((((((((((.....))..)))))).))))).))))))))) (-20.10 = -22.98 + 2.87)

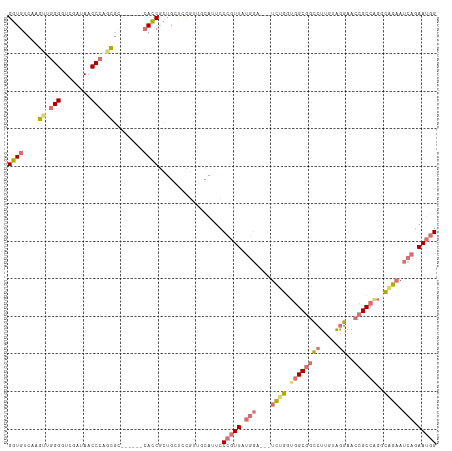
| Location | 19,160,121 – 19,160,230 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.97 |
| Shannon entropy | 0.23146 |
| G+C content | 0.61276 |
| Mean single sequence MFE | -45.83 |
| Consensus MFE | -33.13 |
| Energy contribution | -34.13 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.632635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19160121 109 - 22422827 -ACGCGUAAGGCGGUGAUGGGCGGACGGCACUUCCAUAGCCACUUCCGGUGGCUAUGUUUGGCCCGGCCUUCGAUCGGUGUCAAGUUGGGUUCGAUAACCCAGCGCCACC -..((....((((.((((.((.((.(((..(...(((((((((.....)))))))))...)..))).)).)).)))).))))..((((((((....)))))))))).... ( -48.00, z-score = -1.90, R) >droSec1.super_8 1454429 102 - 3762037 --AACGAAAGGCGGUGCUGGGCGGCCGGCACUUCCACA------UCCGGUGGCUAUGUUUGGCCCGGCCUUCGAUCGGUGUCAAGUUGGGUUCGAUAACCCAGCGCCACC --.......((.((((((((....)))))))).))...------...((((((.....((((((((..(...)..))).)))))((((((((....)))))))))))))) ( -47.80, z-score = -2.34, R) >droEre2.scaffold_4690 9418996 104 - 18748788 AAUGAGAGGGGCAGCGCUGGGCGGCAAGCACUUCCACA------UCCGGUGGCUAUGUUUGGCGCUGCCUUCGAUCGGUGUCAAGUAGGGGUCGAUAACCCAUCGCCAUC ...((..((((((((((..((((...(((....((...------...))..))).))))..)))))))).))..))((((........((((.....))))..))))... ( -41.70, z-score = -1.37, R) >consensus _AAGCGAAAGGCGGUGCUGGGCGGCCGGCACUUCCACA______UCCGGUGGCUAUGUUUGGCCCGGCCUUCGAUCGGUGUCAAGUUGGGUUCGAUAACCCAGCGCCACC .........((.(((((((......))))))).))............((((((.....(((((...(((.......))))))))((((((........)))))))))))) (-33.13 = -34.13 + 1.01)

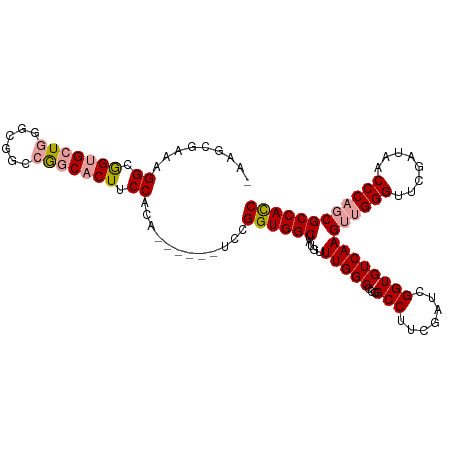
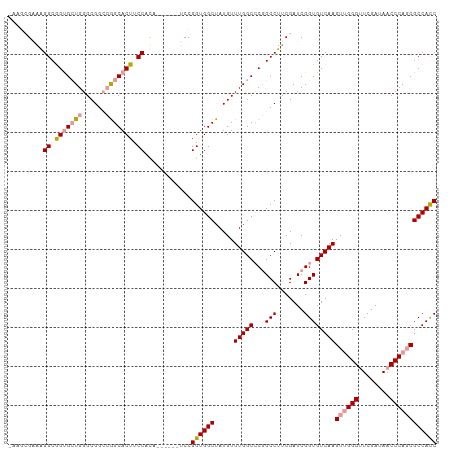
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:44 2011