| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,159,421 – 19,159,551 |
| Length | 130 |
| Max. P | 0.730879 |

| Location | 19,159,421 – 19,159,551 |
|---|---|
| Length | 130 |
| Sequences | 4 |
| Columns | 146 |
| Reading direction | forward |
| Mean pairwise identity | 77.08 |
| Shannon entropy | 0.34911 |
| G+C content | 0.46607 |
| Mean single sequence MFE | -40.80 |
| Consensus MFE | -22.60 |
| Energy contribution | -26.10 |
| Covariance contribution | 3.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.645167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
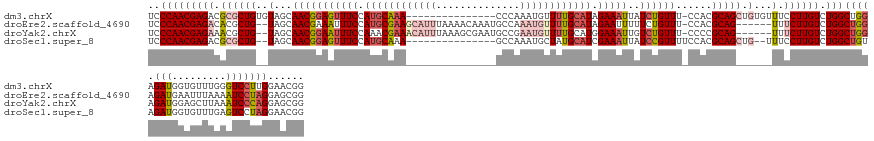
>dm3.chrX 19159421 130 + 22422827 UCCCAACGAGACGCGCUGUGUAGCAACGGAGUUUCCAUGCAAA---------------CCCAAAUGUUUUGCAUAGAAAUUAUCUGUUU-CCACGCAGCUGUGUUUCCUUGUCUGGCUGGAGAUGGUGUUUGGGUCCUUGGAACGG .(((((.((((((((((((((.(.(((((((((((.(((((((---------------.(.....).))))))).)))))..)))))).-).))))))).))))))).(..(((......)))..)...)))))((....)).... ( -44.50, z-score = -2.18, R) >droEre2.scaffold_4690 9418278 137 + 18748788 UCCCAACGAGACACGCUG--UAGCAACGAAAUUUCCAUGCGAAGCAUUUAAAACAAAUGCCAAAUGUUUUGCAUAGAAUUUUUCUGUUU-CCACGCAG------UUUCUUGUCUGGCUGGAGAUGAAUUUAAAAUCCUAGGAGCGG ..(((((((((.((((.(--(.(.(((((((.(((.(((((((((((((............))))))))))))).)))..)))).))).-).)))).)------).)))))).)))((((.(((.........)))))))...... ( -35.70, z-score = -1.75, R) >droYak2.chrX 9604378 137 - 21770863 UCCCAACGAGAAACGCUG--UAGCAACGGAAUUUCCAAACGAAACAUUUAAAGCGAAUGCCGAAUGUUUUGCAUGGAAAUUGUCUGUUU-CCCCGCAG------UUUCUUGUCUGGCUGGAGAUGGAGCUUAAAUCCCAGGAGCGG ..((((((((((((((.(--..(.(((((((((((((..((((((((((...((....)).))))))))))..)))))))..)))))).-)..))).)------)))))))).)))((((.(((.........)))))))...... ( -44.70, z-score = -2.70, R) >droSec1.super_8 1453715 127 + 3762037 UCCCAACGAGACGCGCUG--UAGCAACGGAGUUUCCAUGCAAA---------------GCCAAAUGCUAUGCAUCGAAAUUAUCCGUUUUCCACGCAGCUG--UUUCCUUGUCUGGCUGUAGAUGGUGUUUGAGUCCUAGGAACGG ((((((.((((((.((((--(...(((((((((((.(((((.(---------------((.....))).))))).)))))..))))))......)))))))--)))).)))...((((.(((((...)))))))))...))).... ( -38.30, z-score = -1.29, R) >consensus UCCCAACGAGACACGCUG__UAGCAACGGAAUUUCCAUGCAAA_______________GCCAAAUGUUUUGCAUAGAAAUUAUCUGUUU_CCACGCAG______UUCCUUGUCUGGCUGGAGAUGGAGUUUAAAUCCUAGGAACGG ..((((((((....(((....)))(((((((((((.(((((((((((((............))))))))))))).)))))..))))))...................))))).)))((((.(((.........)))))))...... (-22.60 = -26.10 + 3.50)


| Location | 19,159,421 – 19,159,551 |
|---|---|
| Length | 130 |
| Sequences | 4 |
| Columns | 146 |
| Reading direction | reverse |
| Mean pairwise identity | 77.08 |
| Shannon entropy | 0.34911 |
| G+C content | 0.46607 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -21.03 |
| Energy contribution | -21.78 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.730879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19159421 130 - 22422827 CCGUUCCAAGGACCCAAACACCAUCUCCAGCCAGACAAGGAAACACAGCUGCGUGG-AAACAGAUAAUUUCUAUGCAAAACAUUUGGG---------------UUUGCAUGGAAACUCCGUUGCUACACAGCGCGUCUCGUUGGGA ((.......))..............((((((.((((..(....)...((((.((((-.(((.((...((((((((((((.(....)..---------------)))))))))))).)).))).)))).))))..)))).)))))). ( -43.70, z-score = -3.07, R) >droEre2.scaffold_4690 9418278 137 - 18748788 CCGCUCCUAGGAUUUUAAAUUCAUCUCCAGCCAGACAAGAAA------CUGCGUGG-AAACAGAAAAAUUCUAUGCAAAACAUUUGGCAUUUGUUUUAAAUGCUUCGCAUGGAAAUUUCGUUGCUA--CAGCGUGUCUCGUUGGGA ((.......))..............((((((.(((((.....------.(((((((-.(((.((((..((((((((.((.(((((((........))))))).)).)))))))).))))))).)))--).)))))))).)))))). ( -35.80, z-score = -1.34, R) >droYak2.chrX 9604378 137 + 21770863 CCGCUCCUGGGAUUUAAGCUCCAUCUCCAGCCAGACAAGAAA------CUGCGGGG-AAACAGACAAUUUCCAUGCAAAACAUUCGGCAUUCGCUUUAAAUGUUUCGUUUGGAAAUUCCGUUGCUA--CAGCGUUUCUCGUUGGGA ......(((((((.........))).))))((.(((.(((((------(.(((((.-...)....((((((((.((.(((((((.(((....)))...))))))).)).)))))))))))).((..--..)))))))).))).)). ( -40.30, z-score = -1.61, R) >droSec1.super_8 1453715 127 - 3762037 CCGUUCCUAGGACUCAAACACCAUCUACAGCCAGACAAGGAAA--CAGCUGCGUGGAAAACGGAUAAUUUCGAUGCAUAGCAUUUGGC---------------UUUGCAUGGAAACUCCGUUGCUA--CAGCGCGUCUCGUUGGGA (((((.....)))..............((((.((((..(....--).((.((((((..((((((...((((.(((((.(((.....))---------------).))))).)))).)))))).)))--).)))))))).)))))). ( -43.80, z-score = -2.93, R) >consensus CCGCUCCUAGGACUCAAACACCAUCUCCAGCCAGACAAGAAA______CUGCGUGG_AAACAGAUAAUUUCUAUGCAAAACAUUUGGC_______________UUCGCAUGGAAACUCCGUUGCUA__CAGCGCGUCUCGUUGGGA ((.......))..............((((((.((((..(((.......(((.........)))....(((((((((..............................))))))))).)))((((.....))))..)))).)))))). (-21.03 = -21.78 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:42 2011