| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,114,431 – 19,114,493 |
| Length | 62 |
| Max. P | 0.865891 |

| Location | 19,114,431 – 19,114,493 |
|---|---|
| Length | 62 |
| Sequences | 11 |
| Columns | 65 |
| Reading direction | reverse |
| Mean pairwise identity | 83.43 |
| Shannon entropy | 0.34645 |
| G+C content | 0.48822 |
| Mean single sequence MFE | -18.15 |
| Consensus MFE | -11.90 |
| Energy contribution | -11.65 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.865891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
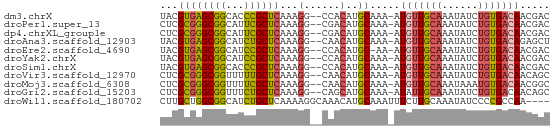
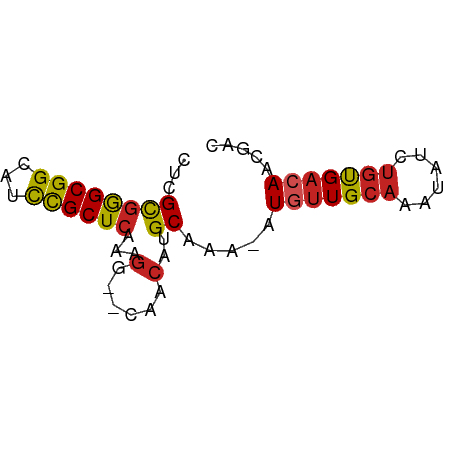
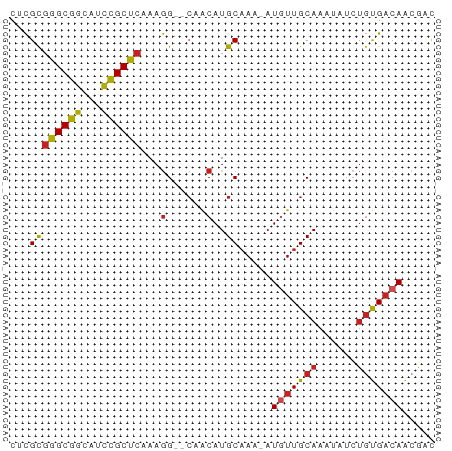
>dm3.chrX 19114431 62 - 22422827 UACGUGAGCGGCACCCGCUCAAAGG--CCACAUGCAAA-AUGUUGCAAAUAUCUGUGACAACGAC ..(((((((((...)))))).....--...........-.(((..((......))..)))))).. ( -17.70, z-score = -1.92, R) >droPer1.super_13 2042933 62 + 2293547 CUCGCGGGCGGCAUUCGCUCAAAGG--CGACAUGCAAA-AUGUUGCAAAUAUCUGUGACAACGAC .(((((((((.....)))))....(--((((((.....-)))))))........))))....... ( -19.50, z-score = -2.23, R) >dp4.chrXL_group1e 8314847 62 - 12523060 CUCGCGGGCGGCAUUCGCUCAAAGG--CGACAUGCAAA-AUGUUGCAAAUAUCUGUGACAACGAC .(((((((((.....)))))....(--((((((.....-)))))))........))))....... ( -19.50, z-score = -2.23, R) >droAna3.scaffold_12903 185109 62 + 802071 UACGUGAGCGGCAUCUGCUCAAAGG--CAACAUGCAAA-AUGUUGCAAAUAUCUGUGACAGAGCU ....(((((((...)))))))...(--((((((.....-))))))).....((((...))))... ( -19.10, z-score = -1.86, R) >droEre2.scaffold_4690 9375641 61 - 18748788 UACGUGAGCGGCAUCCGCUCAAAGG--CCACAUGC-AA-AUGUUGCAAAUAUCUGUGACAACGAC ..(((((((((...)))))).....--........-..-.(((..((......))..)))))).. ( -17.70, z-score = -1.83, R) >droYak2.chrX 9559637 62 + 21770863 UACGUGAGCGGCAUCCGCUCAAAGG--CCACAUGCAAA-AUGUUGCAAAUAUCUGUGACAACGAC ..(((((((((...)))))).....--...........-.(((..((......))..)))))).. ( -17.70, z-score = -1.92, R) >droSim1.chrX 14779737 62 - 17042790 UACGUGAGCGGCACCCGCUCAAAGG--CCACAUGCAAA-AUGUUGCAAAUAUCUGUGACAACGAC ..(((((((((...)))))).....--...........-.(((..((......))..)))))).. ( -17.70, z-score = -1.92, R) >droVir3.scaffold_12970 9366937 62 - 11907090 CUCGCGGGCGGUUUUUGCUCAAAGG--CAACAUGCAAA-AUGUUGCAAAUAUCUGUGACAACAGC .(((((((((.....)))))....(--((((((.....-)))))))........))))....... ( -17.60, z-score = -1.63, R) >droMoj3.scaffold_6308 1987781 62 + 3356042 CUCGCGGGCGGUUUUCGCUCAAAGG--CAACAUGCAAA-AUGUUGCAAAUAAAUGUGACAACGGC .(((((((((.....)))))....(--((((((.....-)))))))........))))....... ( -19.80, z-score = -2.69, R) >droGri2.scaffold_15203 9975311 62 + 11997470 CUCGCGGGCGGUUUCUGCUCAAAGG--CAGCAUGCAAA-AUAUUGCAAAUAUCUGUGACAACAGC .(((((((......(((((....))--)))..(((((.-...)))))....)))))))....... ( -15.70, z-score = -0.82, R) >droWil1.scaffold_180702 2998957 61 - 4511350 CUUGCUGGCGGCAUCUGCUCAAAAGGCAAACAUGCAAAUUUCUUGCAAAUAUCCCCGCCAA---- .....((((((....((((.....))))....(((((.....))))).......)))))).---- ( -17.60, z-score = -2.01, R) >consensus CUCGCGGGCGGCAUCCGCUCAAAGG__CAACAUGCAAA_AUGUUGCAAAUAUCUGUGACAACGAC .....((((((...))))))....................(((((((......)))))))..... (-11.90 = -11.65 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:35 2011