| Sequence ID | dm3.chr2L |
|---|---|
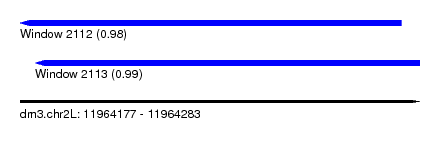
| Location | 11,964,177 – 11,964,283 |
| Length | 106 |
| Max. P | 0.990162 |

| Location | 11,964,177 – 11,964,278 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 88.61 |
| Shannon entropy | 0.20109 |
| G+C content | 0.43381 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -21.54 |
| Energy contribution | -21.98 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
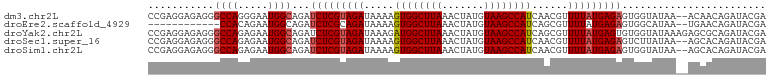
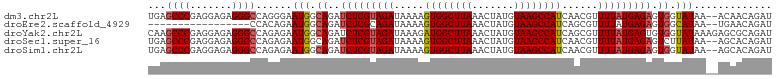
>dm3.chr2L 11964177 101 - 23011544 CCGAGGAGAGGGCCAGGGAAUGGCAGAUCUCGUAGAUAAAAGUGGCUUAAACUAUGUAAGCCAUCAACGUUUUAUGAGAGUGGUAUAA--ACAACAGAUACGA ((.......))((((.....))))...(((((((((.....((((((((.......))))))))......)))))))))...((((..--.......)))).. ( -26.00, z-score = -2.16, R) >droEre2.scaffold_4929 13195318 89 + 26641161 ------------CCACAGAAUGGCAGAUCUCGCAGAUAAAAGUGGCUUAAACUAUGUAAGCCAUCAGCGUUUUAUGAGAGUGGCAUAA--UGAACAGAUACGA ------------...((..(((.((..(((((.(((.....((((((((.......))))))))......))).))))).)).)))..--))........... ( -20.90, z-score = -1.39, R) >droYak2.chr2L 8386236 103 - 22324452 CCGAGGAGAGGGCCAGAGAAUGGCAGAUCUCGUAGAUAAAGAUGGCUUAAACUAUGUAAGCCAUCAGCGUUUUAUGAGUGUGGUAUAAAGAGCGCAGAUACGA .....((((..((((.....))))...))))(((......(((((((((.......))))))))).(((((((...............)))))))...))).. ( -32.76, z-score = -3.75, R) >droSec1.super_16 163556 101 - 1878335 CCGAGGAGAGGGCCAGAGAAUGGCAGAUCUCGUAGAUAAAAGUGGCUUAAACUAUGUAAGCCAUCAACGUUUUAUGAGAGUCUUAUAA--AGCACAGAUACGA .....((((..((((.....))))...))))(((.......((((((((.......))))))))....((((((((((...)))))))--))).....))).. ( -30.40, z-score = -3.64, R) >droSim1.chr2L 11771281 101 - 22036055 CCGAGGAGAGGGCCAGAGAAUGGCAGAUCUCGUAGAUAAAAGUGGCUUAAACUAUGUAAGCCAUCAACGUUUUAUGAGAGUGGUAUAA--AGCACAGAUACGA ((.......))((((.....))))...(((((((((.....((((((((.......))))))))......)))))))))(((.(....--).)))........ ( -27.60, z-score = -2.85, R) >consensus CCGAGGAGAGGGCCAGAGAAUGGCAGAUCUCGUAGAUAAAAGUGGCUUAAACUAUGUAAGCCAUCAACGUUUUAUGAGAGUGGUAUAA__AGCACAGAUACGA ...........((((.....))))...(((((((((.....((((((((.......))))))))......)))))))))........................ (-21.54 = -21.98 + 0.44)
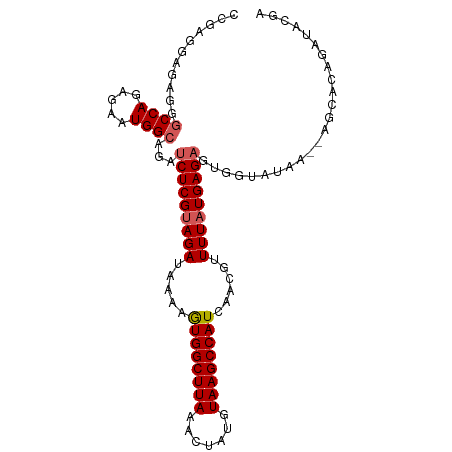
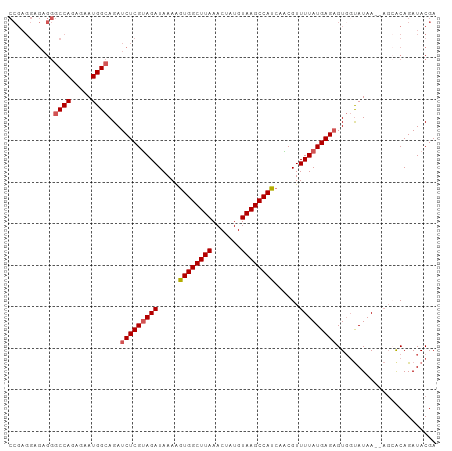
| Location | 11,964,181 – 11,964,283 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 86.19 |
| Shannon entropy | 0.24634 |
| G+C content | 0.43741 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -23.09 |
| Energy contribution | -23.72 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.990162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
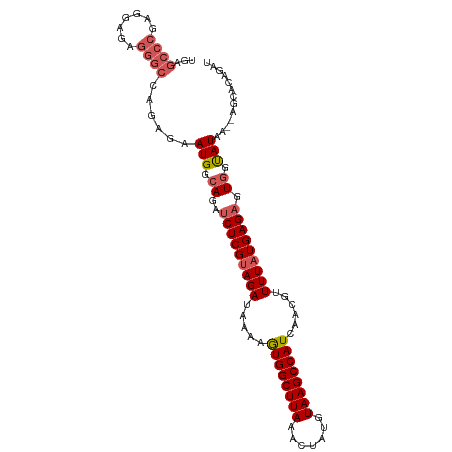
>dm3.chr2L 11964181 102 - 23011544 UGAGCCCGAGGAGAGGGCCAGGGAAUGGCAGAUCUCGUAGAUAAAAGUGGCUUAAACUAUGUAAGCCAUCAACGUUUUAUGAGAGUGGUAUAA--ACAACAGAU ((.((((.......))))))....(((.((..(((((((((.....((((((((.......))))))))......))))))))).)).)))..--......... ( -29.60, z-score = -2.89, R) >droEre2.scaffold_4929 13195322 85 + 26641161 -----------------CCACAGAAUGGCAGAUCUCGCAGAUAAAAGUGGCUUAAACUAUGUAAGCCAUCAGCGUUUUAUGAGAGUGGCAUAA--UGAACAGAU -----------------...((..(((.((..(((((.(((.....((((((((.......))))))))......))).))))).)).)))..--))....... ( -20.90, z-score = -1.67, R) >droYak2.chr2L 8386240 104 - 22324452 CAAGCCCGAGGAGAGGGCCAGAGAAUGGCAGAUCUCGUAGAUAAAGAUGGCUUAAACUAUGUAAGCCAUCAGCGUUUUAUGAGUGUGGUAUAAAGAGCGCAGAU ..........((((..((((.....))))...)))).........(((((((((.......))))))))).(((((((...............))))))).... ( -31.26, z-score = -2.94, R) >droSec1.super_16 163560 102 - 1878335 UGAGCCCGAGGAGAGGGCCAGAGAAUGGCAGAUCUCGUAGAUAAAAGUGGCUUAAACUAUGUAAGCCAUCAACGUUUUAUGAGAGUCUUAUAA--AGCACAGAU ((.((.....((((..((((.....))))...(((((((((.....((((((((.......))))))))......))))))))).))))....--.)).))... ( -29.60, z-score = -2.95, R) >droSim1.chr2L 11771285 102 - 22036055 UGAGCCCGAGGAGAGGGCCAGAGAAUGGCAGAUCUCGUAGAUAAAAGUGGCUUAAACUAUGUAAGCCAUCAACGUUUUAUGAGAGUGGUAUAA--AGCACAGAU ((.((((.......))))))....(((.((..(((((((((.....((((((((.......))))))))......))))))))).)).)))..--......... ( -29.60, z-score = -3.14, R) >consensus UGAGCCCGAGGAGAGGGCCAGAGAAUGGCAGAUCUCGUAGAUAAAAGUGGCUUAAACUAUGUAAGCCAUCAACGUUUUAUGAGAGUGGUAUAA__AGCACAGAU ...((((.......))))......(((.((..(((((((((.....((((((((.......))))))))......))))))))).)).)))............. (-23.09 = -23.72 + 0.63)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:34:02 2011