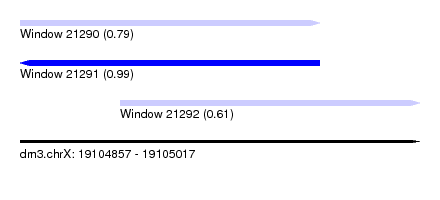
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,104,857 – 19,105,017 |
| Length | 160 |
| Max. P | 0.990307 |

| Location | 19,104,857 – 19,104,977 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.21 |
| Shannon entropy | 0.40028 |
| G+C content | 0.56264 |
| Mean single sequence MFE | -42.04 |
| Consensus MFE | -19.92 |
| Energy contribution | -20.12 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.791751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
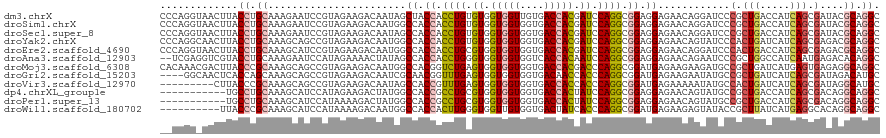
>dm3.chrX 19104857 120 + 22422827 CCCAGGUAACUUACCUGCAAAGAAUCCGUAGAAGACAAUAGCUACCACCUGUGUGGUGGUUGUGACCACGAUCCAGGCGGAGGAGAACAGGAUCCCGCUGACCAUCAGCGAUACGCAGGC .............(((((.....((((((.....)).....((.((.((((..(.(((((....))))).)..)))).)).))......))))..(((((.....)))))....))))). ( -40.00, z-score = -1.91, R) >droSim1.chrX 14769662 120 + 17042790 CCCAGGUAACUUACCUGCAAAGAAUCCGUAGAAGACAAUGGCCACCACCUGUGUGGUGGUGGUGACCACGAUCCAGGCGGAGGAGAACAGGAUCCCGCUGACCAUCAGCGAUACGCAGGC .............(((((......(((((.((......(.(((((((((.....))))))))).)......))...)))))(((........)))(((((.....)))))....))))). ( -43.90, z-score = -2.03, R) >droSec1.super_8 1399644 120 + 3762037 CCCAGGUAACUUACCUGCAAAGAAUCCGUAGAAGACAAUAGCCACCACCUGUGUGGUGGUGGUGACCACGAUCCAGGCGGAGGAGAACAGGAUCCCGCUGACCAUCAGCGAUACGCAGGC .............(((((......(((((.((........(((((((((.....)))))))))........))...)))))(((........)))(((((.....)))))....))))). ( -44.29, z-score = -2.68, R) >droYak2.chrX 17694205 120 + 21770863 CCCAGGCAACUUACCUGCAAAGCAGCCGUAGAAGACAAUGGCCACCACCUGUGUGGUGGUGGUGACCACGAUCCAGGCGGAGGAGAACAGUAUCCCACUGAUCAUCAGCGAGACGCAGGC ...((....))..(((((...((.(((((........)))))..((.((((..(.(((((....))))).)..)))).))..((((.((((.....)))).)).)).)).....))))). ( -40.60, z-score = -0.87, R) >droEre2.scaffold_4690 9366024 120 + 18748788 CCCAGGUAACUUACCUGCAAAGCAUCCGUAGAAGACAAUGGCCACCACCUGCGUGGUGGUGGUGACCACGAUCCAGGCGGAGGAGAACAGGAUCCCACUGACCAUCAGCGAGACGCAGGC ((((((((...))))))....((.(((((.((......(.(((((((((.....))))))))).)..........(((((.((.((......)))).))).)).)).))).)).)).)). ( -40.00, z-score = -0.76, R) >droAna3.scaffold_12903 174978 118 - 802071 --UCGAGGUCGUACCUGCAAAGAAUCCAUAGAAAACUAUAGCCACCACCUGGGUGGUGGUGGUCACCACAAUCCAGGCGGAGGAGAACAGAAUCCCGCUGGCCAUCAAUGAGACACAGGC --...........((((..........((((....)))).((((((.(((((((.(((((....))))).))))))).)).(((........)))...)))).............)))). ( -36.00, z-score = -0.57, R) >droMoj3.scaffold_6308 1975403 120 - 3356042 CACAAACGACUUACCCGCAAAGCAGCCGUAGAAGACAAUGGCCACGGUCUGAGUGGUGGUGGUGACCACGACCCAGGCGGAUGAGAAGAAGAUGCCGCUGAUCAUGAGUGAGAGGCAGGC ......(..((((.((((......(((((........)))))......(((.((.(((((....))))).)).))))))).))))..).....((((((..(((....)))..))).))) ( -39.90, z-score = -1.78, R) >droGri2.scaffold_15203 9965168 116 - 11997470 ----GGCAACUCACCAGCAAAGCAGCCGUAGAAGACAAUCGCAACGGUUUGAGUGGUGGUGGUGACAACCACCCAGGCGGAUGAGAAGAAUAUGCCGCUGAUCAUCAGCGAUAGACAUGC ----.(((.((((((.((.....((((((.((......))...))))))...(.(((((((....).)))))))..)))).))))..........(((((.....))))).......))) ( -36.90, z-score = -1.70, R) >droVir3.scaffold_12970 9352888 111 + 11907090 ---------CUUACCCGCAAAGCAGCCGUAGAAGACAAUAGCCACCGUUUGAGUGGUGGUGGUGACCACCACCCAGGCGGAUGAGAAAAAUAUGCCACUGAUCAUCAGCGAUAGGCAUGC ---------............((((((...............(((((((((.((((((((....)))))))).))))))).))............(.(((.....))).)...))).))) ( -36.00, z-score = -2.31, R) >dp4.chrXL_group1e 8304946 109 + 12523060 -----------UGCCUGCAAAGCAUCCAUAGAAGACUAUGGCCACCGCCUGCGUGGUGGUGGUGACCACUAUCCAGGCGGAGGAGAACAGUAUGCCGCUGACCAUCAGCGACAGGCAGGC -----------.((((((...((((((((((....))))))((.(((((((.((((((((....)))))))).))))))).))........))))(((((.....)))))....)))))) ( -57.10, z-score = -5.68, R) >droPer1.super_13 2032363 109 - 2293547 -----------UGCCUGCAAAGCAUCCAUAAAAGACUAUGGCCACCGCCUGCGUGGUGGUGGUGACCACUAUCCAGGCGGAGGAGAACAGUAUGCCGCUGACCAUCAGCGACAGGCAGGC -----------.((((((...(((((((((......)))))((.(((((((.((((((((....)))))))).))))))).))........))))(((((.....)))))....)))))) ( -54.60, z-score = -5.19, R) >droWil1.scaffold_180702 2991440 110 + 4511350 ----------UUACCCGCAAAGCAUCCAUAAAAGACAAUGGCCACCACUUGGGUGGUUGUGGUGACUAUCACCCAGGCGGAUGAGAAGAGUAUACCGCUUAUCAUGAGGCACAGGCAGGC ----------...((.((........(((........)))(((.((.((((((((((...(....).)))))))))).))((((...((((.....)))).))))..)))....)).)). ( -35.20, z-score = -1.35, R) >consensus __CAGG_AACUUACCUGCAAAGCAUCCGUAGAAGACAAUGGCCACCACCUGUGUGGUGGUGGUGACCACGAUCCAGGCGGAGGAGAACAGUAUCCCGCUGACCAUCAGCGAUACGCAGGC .............((.((.......................((.((.((((.((.(((((....))))).)).)))).)).))............(.(((.....))).)....)).)). (-19.92 = -20.12 + 0.20)
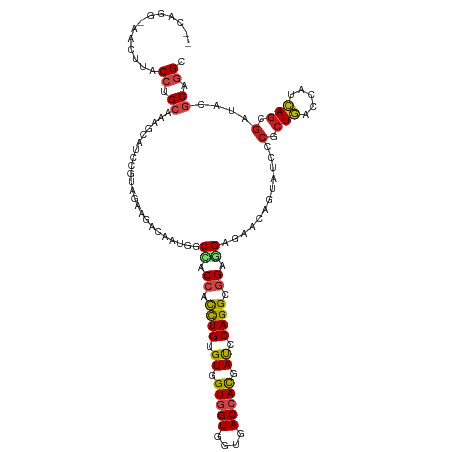
| Location | 19,104,857 – 19,104,977 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.21 |
| Shannon entropy | 0.40028 |
| G+C content | 0.56264 |
| Mean single sequence MFE | -46.74 |
| Consensus MFE | -26.51 |
| Energy contribution | -25.96 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.990307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
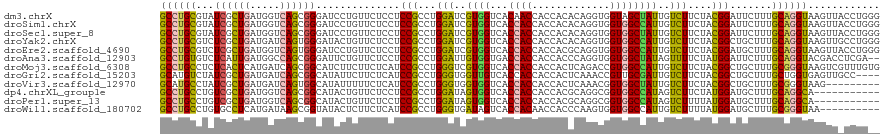
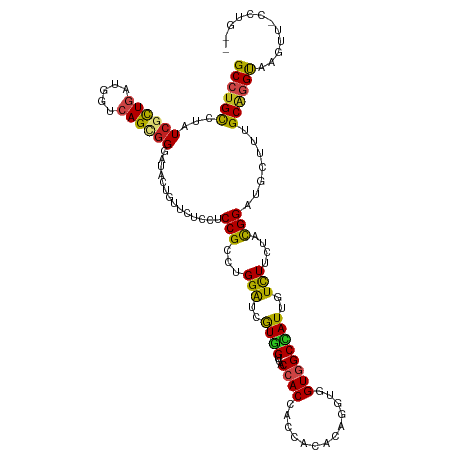
>dm3.chrX 19104857 120 - 22422827 GCCUGCGUAUCGCUGAUGGUCAGCGGGAUCCUGUUCUCCUCCGCCUGGAUCGUGGUCACAACCACCACACAGGUGGUAGCUAUUGUCUUCUACGGAUUCUUUGCAGGUAAGUUACCUGGG (((((((...(((((.....)))))((((((.......(((((((((..(.(((((....))))).)..))))))).)).....((.....))))))))..)))))))............ ( -44.10, z-score = -1.90, R) >droSim1.chrX 14769662 120 - 17042790 GCCUGCGUAUCGCUGAUGGUCAGCGGGAUCCUGUUCUCCUCCGCCUGGAUCGUGGUCACCACCACCACACAGGUGGUGGCCAUUGUCUUCUACGGAUUCUUUGCAGGUAAGUUACCUGGG (((((((...(((((.....)))))(((........)))((((...((((.((((((((((((........)))))))))))).))))....)))).....)))))))............ ( -51.50, z-score = -3.18, R) >droSec1.super_8 1399644 120 - 3762037 GCCUGCGUAUCGCUGAUGGUCAGCGGGAUCCUGUUCUCCUCCGCCUGGAUCGUGGUCACCACCACCACACAGGUGGUGGCUAUUGUCUUCUACGGAUUCUUUGCAGGUAAGUUACCUGGG (((((((...(((((.....)))))(((........)))((((...((((.((((((((((((........)))))))))))).))))....)))).....)))))))............ ( -49.20, z-score = -2.74, R) >droYak2.chrX 17694205 120 - 21770863 GCCUGCGUCUCGCUGAUGAUCAGUGGGAUACUGUUCUCCUCCGCCUGGAUCGUGGUCACCACCACCACACAGGUGGUGGCCAUUGUCUUCUACGGCUGCUUUGCAGGUAAGUUGCCUGGG (((((((((((((((.....))))))))).............(((.((((.((((((((((((........)))))))))))).)))).....)))......))))))............ ( -53.50, z-score = -3.31, R) >droEre2.scaffold_4690 9366024 120 - 18748788 GCCUGCGUCUCGCUGAUGGUCAGUGGGAUCCUGUUCUCCUCCGCCUGGAUCGUGGUCACCACCACCACGCAGGUGGUGGCCAUUGUCUUCUACGGAUGCUUUGCAGGUAAGUUACCUGGG (((((((((((((((.....)))))))))..........((((...((((.((((((((((((........)))))))))))).))))....))))......))))))............ ( -54.30, z-score = -3.34, R) >droAna3.scaffold_12903 174978 118 + 802071 GCCUGUGUCUCAUUGAUGGCCAGCGGGAUUCUGUUCUCCUCCGCCUGGAUUGUGGUGACCACCACCACCCAGGUGGUGGCUAUAGUUUUCUAUGGAUUCUUUGCAGGUACGACCUCGA-- (((((((((.....)))..(((..((((..((((...((.((((((((...((((((.....)))))))))))))).))..))))..)))).))).......)))))).((....)).-- ( -47.50, z-score = -2.87, R) >droMoj3.scaffold_6308 1975403 120 + 3356042 GCCUGCCUCUCACUCAUGAUCAGCGGCAUCUUCUUCUCAUCCGCCUGGGUCGUGGUCACCACCACCACUCAGACCGUGGCCAUUGUCUUCUACGGCUGCUUUGCGGGUAAGUCGUUUGUG (((.((...(((....)))...)))))........((.((((((((((((.(((((....))))).))))))...(..(((............)))..)...)))))).))......... ( -38.00, z-score = -1.63, R) >droGri2.scaffold_15203 9965168 116 + 11997470 GCAUGUCUAUCGCUGAUGAUCAGCGGCAUAUUCUUCUCAUCCGCCUGGGUGGUUGUCACCACCACCACUCAAACCGUUGCGAUUGUCUUCUACGGCUGCUUUGCUGGUGAGUUGCC---- (((......((((((.....)))))).........((((((.((.((((((((.((....)).))))))))..((((...((......)).)))).......)).)))))).))).---- ( -35.40, z-score = -1.08, R) >droVir3.scaffold_12970 9352888 111 - 11907090 GCAUGCCUAUCGCUGAUGAUCAGUGGCAUAUUUUUCUCAUCCGCCUGGGUGGUGGUCACCACCACCACUCAAACGGUGGCUAUUGUCUUCUACGGCUGCUUUGCGGGUAAG--------- ..(((((....((((.....))))))))).........((((((.(((((((((((....)))))))))))...((..((..(........)..))..))..))))))...--------- ( -42.10, z-score = -3.09, R) >dp4.chrXL_group1e 8304946 109 - 12523060 GCCUGCCUGUCGCUGAUGGUCAGCGGCAUACUGUUCUCCUCCGCCUGGAUAGUGGUCACCACCACCACGCAGGCGGUGGCCAUAGUCUUCUAUGGAUGCUUUGCAGGCA----------- ((((((.((((((((.....)))))))).........((.(((((((....(((((....)))))....))))))).))((((((....)))))).......)))))).----------- ( -54.40, z-score = -4.25, R) >droPer1.super_13 2032363 109 + 2293547 GCCUGCCUGUCGCUGAUGGUCAGCGGCAUACUGUUCUCCUCCGCCUGGAUAGUGGUCACCACCACCACGCAGGCGGUGGCCAUAGUCUUUUAUGGAUGCUUUGCAGGCA----------- ((((((.((((((((.....)))))))).........((.(((((((....(((((....)))))....))))))).))((((((....)))))).......)))))).----------- ( -52.50, z-score = -3.92, R) >droWil1.scaffold_180702 2991440 110 - 4511350 GCCUGCCUGUGCCUCAUGAUAAGCGGUAUACUCUUCUCAUCCGCCUGGGUGAUAGUCACCACAACCACCCAAGUGGUGGCCAUUGUCUUUUAUGGAUGCUUUGCGGGUAA---------- ((((((.((((((.(.......).)))))).......((((((...(((..((.((((((((..........)))))))).))..)))....))))))....))))))..---------- ( -38.40, z-score = -2.02, R) >consensus GCCUGCCUAUCGCUGAUGGUCAGCGGGAUACUGUUCUCCUCCGCCUGGAUCGUGGUCACCACCACCACACAGGUGGUGGCCAUUGUCUUCUACGGAUGCUUUGCAGGUAAGUU_CCUG__ ((((((...((((((.....))))))..............(((...(((..((((...((((.............))))))))..)))....))).......))))))............ (-26.51 = -25.96 + -0.54)
| Location | 19,104,897 – 19,105,017 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.37 |
| Shannon entropy | 0.38063 |
| G+C content | 0.59097 |
| Mean single sequence MFE | -48.94 |
| Consensus MFE | -23.87 |
| Energy contribution | -23.58 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.613068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
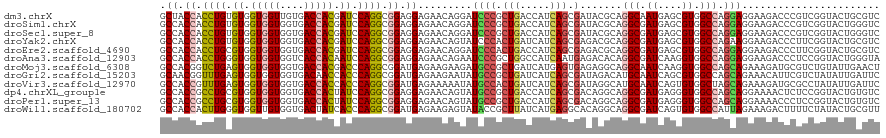
>dm3.chrX 19104897 120 + 22422827 GCUACCACCUGUGUGGUGGUUGUGACCACGAUCCAGGCGGAGGAGAACAGGAUCCCGCUGACCAUCAGCGAUACGCAGGCAAUGAGCGUGGCCAGGAGGAAGACCCGUCGGUACUGCGUC ....((.((((..(.(((((....))))).)..)))).)).(((........)))(((((.....)))))..((((((((.....))...(((.(..((.....))..)))).)))))). ( -44.80, z-score = -0.50, R) >droSim1.chrX 14769702 120 + 17042790 GCCACCACCUGUGUGGUGGUGGUGACCACGAUCCAGGCGGAGGAGAACAGGAUCCCGCUGACCAUCAGCGAUACGCAGGCGAUGAGCGUGGCCAGGAGGAAGACCCGUCGGUACUGGGUC (((((((((.....)))))))))......((((((((((..(((........)))(((((.....)))))...))).(((.((....)).))).(..((.....))..)....))))))) ( -51.30, z-score = -1.44, R) >droSec1.super_8 1399684 120 + 3762037 GCCACCACCUGUGUGGUGGUGGUGACCACGAUCCAGGCGGAGGAGAACAGGAUCCCGCUGACCAUCAGCGAUACGCAGGCGAUGAGCGUGGCCAGGAGGAAGACCCGUCGGUACUGGGUC (((((((((.....)))))))))......((((((((((..(((........)))(((((.....)))))...))).(((.((....)).))).(..((.....))..)....))))))) ( -51.30, z-score = -1.44, R) >droYak2.chrX 17694245 120 + 21770863 GCCACCACCUGUGUGGUGGUGGUGACCACGAUCCAGGCGGAGGAGAACAGUAUCCCACUGAUCAUCAGCGAGACGCAGGCGAUGAGCGUGGCCAGAAGGAAGACCCUUCGGUACUGCGUC .((.((.((((..(.(((((....))))).)..)))).)).)).((.((((.....)))).))........(((((((((.....))...(((.(((((.....)))))))).))))))) ( -49.40, z-score = -2.04, R) >droEre2.scaffold_4690 9366064 120 + 18748788 GCCACCACCUGCGUGGUGGUGGUGACCACGAUCCAGGCGGAGGAGAACAGGAUCCCACUGACCAUCAGCGAGACGCAGGCGAUGAGCGUGGCCAGGAGGAAGACCCUUCGGUACUGCGUC (((((((((.....))))))))).....((.....(((((.((.((......)))).))).)).....)).(((((((((.....))...(((.(((((.....)))))))).))))))) ( -49.10, z-score = -1.20, R) >droAna3.scaffold_12903 175016 120 - 802071 GCCACCACCUGGGUGGUGGUGGUCACCACAAUCCAGGCGGAGGAGAACAGAAUCCCGCUGGCCAUCAAUGAGACACAGGCGAUCAAGGUGGCCAGGAGGAAGACCCUCCGGUACUGGGUA (((((((((.....))))))))).......((((((.((((((.(.......(((..(((((((((..(((...........))).)))))))))..)))...)))))))...)))))). ( -53.90, z-score = -2.27, R) >droMoj3.scaffold_6308 1975443 120 - 3356042 GCCACGGUCUGAGUGGUGGUGGUGACCACGACCCAGGCGGAUGAGAAGAAGAUGCCGCUGAUCAUGAGUGAGAGGCAGGCAAUCAAGGUGGCCAGCAGAAAGAUGCGUCUGUAUUGAACU (((((.(((((.((.(((((....))))).)).)))))............(((((((((..(((....)))..))).))).)))...)))))..(((((........)))))........ ( -44.40, z-score = -2.28, R) >droGri2.scaffold_15203 9965204 120 - 11997470 GCAACGGUUUGAGUGGUGGUGGUGACAACCACCCAGGCGGAUGAGAAGAAUAUGCCGCUGAUCAUCAGCGAUAGACAUGCAAUCAGCGUGGCCAGCAGAAACAUUCGUCUAUAUUGAUUC ....((((....(.(((((((....).)))))))(((((((((.........((((((((.....))))).....(((((.....)))))....)))....)))))))))..)))).... ( -38.62, z-score = -1.57, R) >droVir3.scaffold_12970 9352919 120 + 11907090 GCCACCGUUUGAGUGGUGGUGGUGACCACCACCCAGGCGGAUGAGAAAAAUAUGCCACUGAUCAUCAGCGAUAGGCAUGCAAUCAGUGUGGCUAGCAGAAAGAUGCGCCUAUAUUGAUUC (((((((((((.((((((((....)))))))).))))))).))........(((((.(((.....))).....)))))))((((((((((((..(((......)))))).))))))))). ( -48.00, z-score = -3.98, R) >dp4.chrXL_group1e 8304975 120 + 12523060 GCCACCGCCUGCGUGGUGGUGGUGACCACUAUCCAGGCGGAGGAGAACAGUAUGCCGCUGACCAUCAGCGACAGGCAGGCGAUGAGGGUGGCCAGCAGGAAAACUCUCCGGUACUGUGUC .((.(((((((.((((((((....)))))))).))))))).))...((((((((((((((.....))))(.....).)))...((((((..((....))...))))))..)))))))... ( -55.90, z-score = -2.62, R) >droPer1.super_13 2032392 120 - 2293547 GCCACCGCCUGCGUGGUGGUGGUGACCACUAUCCAGGCGGAGGAGAACAGUAUGCCGCUGACCAUCAGCGACAGGCAGGCGAUGAGGGUGGCCAGCAGGAAAACCCUCCGGUACUGUGUC .((.(((((((.((((((((....)))))))).))))))).))...((((((((((((((.....))))(.....).)))...((((((..((....))...))))))..)))))))... ( -58.90, z-score = -3.31, R) >droWil1.scaffold_180702 2991470 120 + 4511350 GCCACCACUUGGGUGGUUGUGGUGACUAUCACCCAGGCGGAUGAGAAGAGUAUACCGCUUAUCAUGAGGCACAGGCAGGCGAUCAGUGUGGCCAUUAGAAAGACUUUUCUAUACUGCGUU (((.((.((((((((((...(....).)))))))))).))((((...((((.....)))).))))..)))....((((((.((....)).)))..((((((....))))))...)))... ( -41.70, z-score = -1.77, R) >consensus GCCACCACCUGUGUGGUGGUGGUGACCACGAUCCAGGCGGAGGAGAACAGUAUCCCGCUGACCAUCAGCGAUACGCAGGCGAUGAGCGUGGCCAGCAGGAAGACCCGUCGGUACUGCGUC .((.((.((((.((.(((((....))))).)).)))).)).)).........((((.(((.....))).).......(((.((....)).))).)))....................... (-23.87 = -23.58 + -0.29)
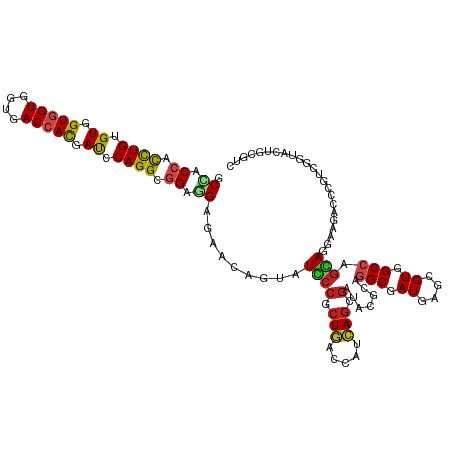
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:33 2011