
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,104,279 – 19,104,390 |
| Length | 111 |
| Max. P | 0.554107 |

| Location | 19,104,279 – 19,104,390 |
|---|---|
| Length | 111 |
| Sequences | 13 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.58 |
| Shannon entropy | 0.61606 |
| G+C content | 0.54515 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -15.91 |
| Energy contribution | -15.94 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
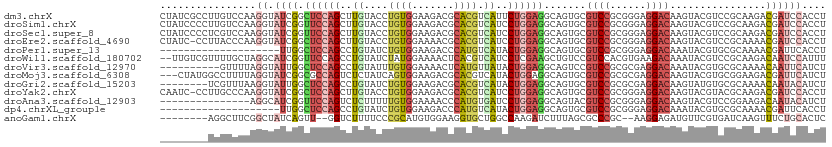
>dm3.chrX 19104279 111 - 22422827 CUAUCGCCUUGUCCAAGGUAUCGGCUCCAGCUUGUACCUGUGGAAGACGCACGUCAUUCUGGAGGCAGUGCGUCCGCGGGAGGACAAGUACGUCCGCAAGACGAUCCACCU ..((((.(((((((.((((((.(((....))).))))))(((((...((((((((........))).))))))))))))..((((......))))))))).))))...... ( -43.10, z-score = -1.82, R) >droSim1.chrX 14769117 111 - 17042790 CUAUCCCCUUGUCCAAGGUAUCGGUUCCAGCUUGUACCUGUGGAAGACGCACGUCAUCCUGGAGGCAGUGCGUCCGCGGGAGGACAAGUACGUCCGCAAGACGAUCCACCU ......((((....))))....((.((...(((((.((((((((...((((((((........))).))))))))))))).((((......)))))))))..)).)).... ( -40.60, z-score = -1.42, R) >droSec1.super_8 1399106 111 - 3762037 CUAUCCCCUCGUCCAAGGUAUCGGUUCCAGCUUGUACCUGUGGAAGACGCACGUCAUCCUGGAGGCAGUGCGUCCGCGGGAGGACAAGUACGUCCGCAAGACGAUCCACCU .....(((.((((((((((((.(((....))).)))))).)))).((((((((((........))).))))))))).))).((((......))))................ ( -40.40, z-score = -1.30, R) >droEre2.scaffold_4690 9365511 110 - 18748788 CUAUC-CCUUACCCAAGGUAUCGGCUCCAGCUUGUACCUGUGGAAAACGCACGUCAUCCUGGAGGCAGUGCGUCCGCGGGAGGACAAGUACGUCCGCAAAACGAUCCACCU ...((-((.......((((((.(((....))).))))))(((((...((((((((........))).))))))))))))))((((......))))................ ( -39.30, z-score = -2.09, R) >droPer1.super_13 2032272 91 + 2293547 --------------------UUGGCUCCAGCCUGUAUCUGUGGAAGACCCAUGUCAUACUGGAGGCAGUGCGUCCGCGGGAGGACAAAUACGUGCGCAAAACGAUUCACCU --------------------.((.((((((..((....(((((.....))))).))..)))))).))(((.((((......)))).....(((.......)))...))).. ( -25.90, z-score = 0.37, R) >droWil1.scaffold_180702 2991279 109 - 4511350 --UUGUCGUUUUGCUAGGCAUCGGUUCCAGCCUGUAUCUAUGGAAAACUCACGUCAUCCUCGAAGCUGUCCGUCCACGUGAAGACAAAUACGUCCGCAAGACAAUCCAUUU --((((((((((.((((..((.(((....))).))..))).).)))))((((((......((........))...)))))).)))))....((.(....)))......... ( -24.50, z-score = -0.64, R) >droVir3.scaffold_12970 9352737 101 - 11907090 ----------GUUUUAGGUAUUGGCUCCAGCCUGUAUUUGUGGAAAACUCAUGUUAUACUGGAGGCAGUCCGUCCGCGCGAGGACAAAUACGUGCGCAAAACAAUUCAUCU ----------(((((.((.((((.((((((..((((..(((((.....))))).)))))))))).))))))....(((((.(........).))))))))))......... ( -30.10, z-score = -1.70, R) >droMoj3.scaffold_6308 1975252 108 + 3356042 ---CUAUGGCCUUUUAGGUAUCGGCGCCAGUCUCUAUCAGUGGAAGACGCACGUCAUACUGGAGGCAGUGCGUCCGCGCGAGGACAAGUACGUGCGGAAGACGAUUCAUCU ---.....(((((...(((((.((((...(((((((....))).))))...))))))))).))))).((.(.((((((((..........)))))))).)))......... ( -36.70, z-score = -0.69, R) >droGri2.scaffold_15203 9964915 103 + 11997470 --------UCGUUUAAGGUAUUGGCUCCAGCCUGUAUCUGUGGAAGACGCACGUCAUACUGGAGGCAGUGCGUCCGCGCGAGGACAAGUAUGUGCGCAAAACAAUACAUCU --------..((((...((((((.((((((..((.((.((((.....)))).))))..)))))).))))))....(((((...(....)...))))).))))......... ( -34.20, z-score = -1.29, R) >droYak2.chrX 17693575 110 - 21770863 CAAUC-CCUUGCCCAAGGUAUCGGCUCCAGCUUGUACCUGUGGAAGACGCACGUCAUCCUGGAGGCAGUGCGUCCGCGGGAGGACAAGUACGUACGCAAGACGAUCCACCU ...((-(((((((((((((((.(((....))).)))))).)))..((((((((((........))).))))))).))))).)))......(((.(....))))........ ( -42.10, z-score = -1.99, R) >droAna3.scaffold_12903 174833 96 + 802071 ---------------AGGCAUCGGUUCCAGUCUCUUUUUGUGGAAAACCCAUGUGAUCCUGGAGGCAGUACGUCCGCGGGAGGACAAGUACGUCCGGAAGACAAUACAUCU ---------------.(((...(.((((((..((....(((((.....))))).))..)))))).).((((((((......))))..)))))))(....)........... ( -28.00, z-score = -0.13, R) >dp4.chrXL_group1e 8304855 91 - 12523060 --------------------UUGGCUCCAGCCUGUAUCUGUGGAAGACCCAUGUCAUACUGGAGGCAGUGCGUCCGCGGGAGGACAAAUACGUGCGCAAAACGAUUCACCU --------------------.((.((((((..((....(((((.....))))).))..)))))).))(((.((((......)))).....(((.......)))...))).. ( -25.90, z-score = 0.37, R) >anoGam1.chrX 13156242 99 - 22145176 --------AGGCUUCGGCUAUCAGUU--GGUCUUUUCCCGCAUGUGGAAGGUGCUGGCCAAGAUCUUUAGCGCCCGC--AAGGAGAUGUUCGUGAUCAAGUUUCUGCACUC --------.(((....)))....((.--((......)).)).((..((((((....)))..((((...(((((((..--..)).).))))...))))....)))..))... ( -27.70, z-score = 0.59, R) >consensus ________UUGUCCAAGGUAUCGGCUCCAGCCUGUAUCUGUGGAAGACGCACGUCAUCCUGGAGGCAGUGCGUCCGCGGGAGGACAAGUACGUCCGCAAGACGAUCCACCU ......................(.((((((..((....((((.......)))).))..)))))).).....((((......)))).......................... (-15.91 = -15.94 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:31 2011