| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,101,727 – 19,101,879 |
| Length | 152 |
| Max. P | 0.798576 |

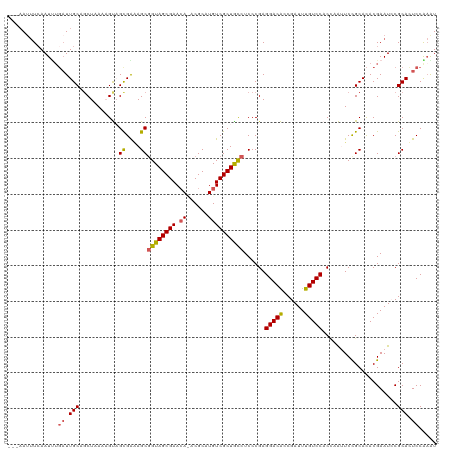
| Location | 19,101,727 – 19,101,840 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.64 |
| Shannon entropy | 0.28121 |
| G+C content | 0.50330 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -23.86 |
| Energy contribution | -23.78 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.798576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
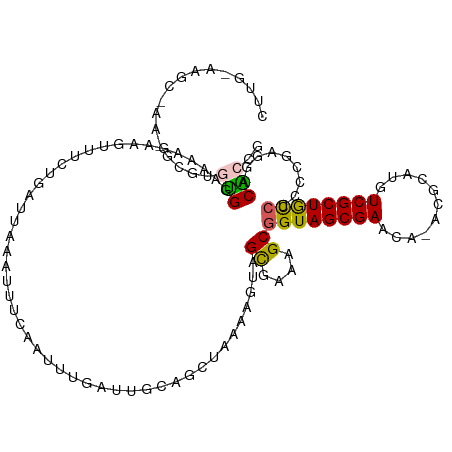
>dm3.chrX 19101727 113 - 22422827 ---AAUUUCAAU----UUGCAGCUAAAAGUAGCGAAAGCGGUAGCGAACACACGCAUGUCGCUGCCUCCCGAGGACACCGAUUUGUCCACAAAUUCCGCAUACGAAUCCGCACUCCAAAU ---.........----.(((.((((....))))....((((((((((.((......))))))))))......(((((......))))).........))..........)))........ ( -29.70, z-score = -2.21, R) >droSim1.chrX 14766568 117 - 17042790 ---AAUUUCAAUUUGAUUGCAGCUAAACGUUGUGAUAGCGGUAGCGAACAUACGCAUGUCGCUACCUCCCGAGGACACCGAUUUGUCCACUAAUUCCGCACACGAAUACGCACUCCAAAU ---.......(((((((..((((.....))))..)).((((((((((.(((....)))))))))))......(((((......))))).........))...)))))............. ( -33.10, z-score = -3.31, R) >droSec1.super_8 1396632 117 - 3762037 ---AAUUUCAAUUUGAUUGCAGCUAAACGUUGUGAGAGCUGUAGCGAACAUACGCAUGUCGCUACCUCCCGAGGACACCGAUUUGUCCACUAAUUCCGCACACGAAUACGCACUCCAAAU ---.......(((((...(((((.....)))))(((.((.(((((((.(((....)))))))))).......(((((......))))).....................)).)))))))) ( -28.30, z-score = -1.95, R) >droYak2.chrX 17691165 118 - 21770863 UCGCAUCUGAAUUUGAUUGCAGCUAAGAGCAGCGGAAGCGGUAGCGAACA--CGCAUGUCGCUGCCUCCCGAGGACGCCGAUUUGUCCACCAGAUCCGCACACGAAUCCGCAUUCCGCGU ..(((((.......)).))).((.....)).((((((((((((((((.((--....))))))))))..........((.((((((.....)))))).))..........)).)))))).. ( -39.60, z-score = -1.11, R) >droEre2.scaffold_4690 9362927 115 - 18748788 ---AAAUUCAAUCUGAUUGCAUCUUACAGCAGCGGAAGCGGUAGCGAAGA--CGCAUGUCGCUGUCUCCCGAGGACGCCGAUUUGUCCACCAGAUCUGCACACGAAUCCGCAUUCCGAGU ---..((((........(((........)))(((((((((..((((....--))).)..))))((((.....))))((.((((((.....)))))).)).......))))).....)))) ( -33.60, z-score = -0.62, R) >consensus ___AAUUUCAAUUUGAUUGCAGCUAAAAGUAGCGAAAGCGGUAGCGAACA_ACGCAUGUCGCUGCCUCCCGAGGACACCGAUUUGUCCACCAAUUCCGCACACGAAUCCGCACUCCAAAU ..............((.(((...........((....))((((((((.((......))))))))))......(((((......))))).....................))).))..... (-23.86 = -23.78 + -0.08)


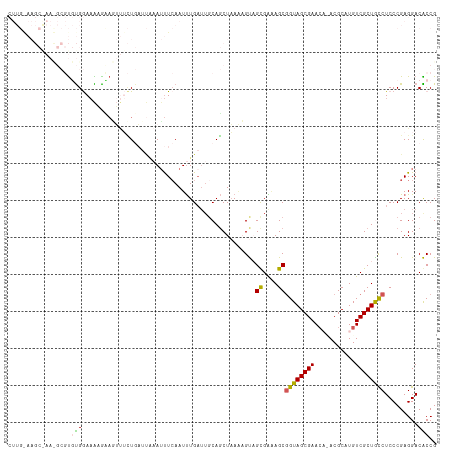
| Location | 19,101,767 – 19,101,879 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 69.61 |
| Shannon entropy | 0.52718 |
| G+C content | 0.48766 |
| Mean single sequence MFE | -31.56 |
| Consensus MFE | -14.40 |
| Energy contribution | -13.96 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.507076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19101767 112 - 22422827 CUUGAAAGCGAAUGCGUGUGGAAAAAUAAUUUCUGAUUAAAUUUCAAUUUG----CAGCUAAAAGUAGCGAAAGCGGUAGCGAACACACGCAUGUCGCUGCCUCCCGAGGACACCG ......((((((((((((((((((..(((((...)))))..))))......----..((((......((....))..))))...))))))))).))))).(((....)))...... ( -35.40, z-score = -2.91, R) >droSim1.chrX 14766608 116 - 17042790 CUUGGAAGCAAACGCGUGUGGAAAAGCAGUUUCUGAUUAAAUUUCAAUUUGAUUGCAGCUAAACGUUGUGAUAGCGGUAGCGAACAUACGCAUGUCGCUACCUCCCGAGGACACCG (((((..((..((((((.(((....((((((..(((.......)))....))))))..))).))).)))....))((((((((.(((....)))))))))))..)))))....... ( -35.00, z-score = -1.96, R) >droSec1.super_8 1396672 116 - 3762037 CUUGGAAGCAAACGCGUGUGGAAAAGCAGUUUCUGAUUAAAUUUCAAUUUGAUUGCAGCUAAACGUUGUGAGAGCUGUAGCGAACAUACGCAUGUCGCUACCUCCCGAGGACACCG (((((.(((..((((((.(((....((((((..(((.......)))....))))))..))).))).)))....)))(((((((.(((....))))))))))...)))))....... ( -31.70, z-score = -0.99, R) >droYak2.chrX 17691205 98 - 21770863 ----------------CUUGAACGCGAUUAAAUUAUUCGCAUCUGAAUUUGAUUGCAGCUAAGAGCAGCGGAAGCGGUAGCGAAC--ACGCAUGUCGCUGCCUCCCGAGGACGCCG ----------------((((...(((((((((((...........))))))))))).((.....)).((....))((((((((.(--(....))))))))))...))))....... ( -31.30, z-score = -1.01, R) >droEre2.scaffold_4690 9362967 94 - 18748788 ----------------UCCGAAAGCGAAUGCAAUA----AAAUUCAAUCUGAUUGCAUCUUACAGCAGCGGAAGCGGUAGCGAAG--ACGCAUGUCGCUGUCUCCCGAGGACGCCG ----------------((((...((..(((((((.----............)))))))......))..))))((((..((((...--.))).)..))))((((.....)))).... ( -24.42, z-score = 0.29, R) >consensus CUUG_AAGC_AA_GCGUGUGGAAAAGAAGUUUCUGAUUAAAUUUCAAUUUGAUUGCAGCUAAAAGUAGCGAAAGCGGUAGCGAACA_ACGCAUGUCGCUGCCUCCCGAGGACACCG .................(((...............................................((....))((((((((...........)))))))).........))).. (-14.40 = -13.96 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:30 2011