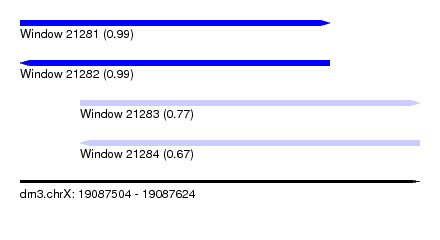
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,087,504 – 19,087,624 |
| Length | 120 |
| Max. P | 0.994759 |

| Location | 19,087,504 – 19,087,597 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 68.89 |
| Shannon entropy | 0.60472 |
| G+C content | 0.49519 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -12.04 |
| Energy contribution | -14.71 |
| Covariance contribution | 2.67 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.994759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
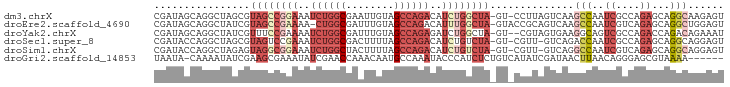
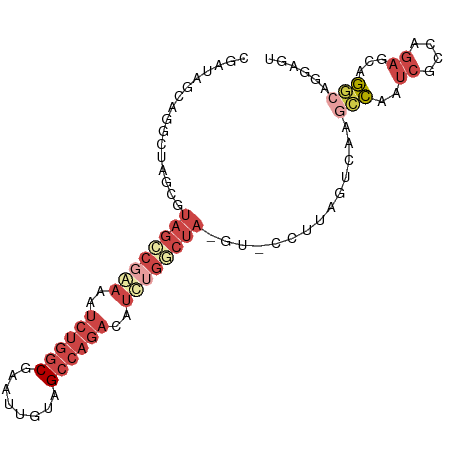
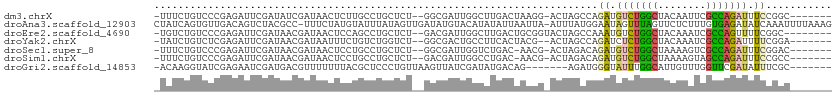
>dm3.chrX 19087504 93 + 22422827 CGAUAGCAGGCUAGCGUAGCCGGAAAUCUGGCGAAUUGUAGCCAGACAUCUGGCUA-GU-CCUUAGUCAAGCCAAUCGCCAGAGCAGGCAAGAGU .(((.((.(((((((.((((((((..((((((........))))))..))))))))-))-...)))))..))..)))(((......)))...... ( -35.80, z-score = -3.10, R) >droEre2.scaffold_4690 9348916 93 + 18748788 CGAUAGCAGGCUAUCGUAGCCGAAAA-CUGGCGAUUUGUAGCCAGACAUUUGGCUA-GUACCGCAGUCAAGCCAAUCGUCAGAGCAGGCUGGAGU ((((((....))))))(((((.....-(((((((((..(((((((....)))))))-.....((......)).)))))))))....))))).... ( -34.10, z-score = -2.79, R) >droYak2.chrX 17676674 92 + 21770863 CGAUAGCAGGCUAUCGUUUCCGAAAAUCUGGCGAUUUGUAGCCAGAGAUCUGGCUA-GU--CGUAGUGAAGGCAGUCGCCAGACCAGACAGAAAU ((((((....))))))((((......((((((((((..(((((((....)))))))-((--(........))))))))))))).......)))). ( -36.42, z-score = -4.43, R) >droSec1.super_8 1381937 92 + 3762037 CGAUACCAGGCUAGCGUAGUCCGAAAUCUGGCGACUUUUAGCCAGACAUCUGUCUA-GU-CGUU-GUCAGACCAAUCGCCAGAGCAGGCAGGAGU .....((..(((.((......(((..(((((((((..((((.(((....))).)))-).-.)))-))))))....))).....)).))).))... ( -28.30, z-score = -1.36, R) >droSim1.chrX 14750149 92 + 17042790 CGAUACCAGGCUAGAGUAGGCGGAAAUCUGGCUACUUUUAGCCAGACAUCUGUCUA-GU-CGUU-GUCAGGCCAAUCGUCAGAGCAGGCAGGAGU .....((.(((..((.((((((((..((((((((....))))))))..))))))))-.)-)...-)))..(((..((....))...))).))... ( -38.00, z-score = -4.13, R) >droGri2.scaffold_14853 2231539 88 - 10151454 UAAUA-CAAAAUAUCGAAGCGAAAUAUCGAACCAAACAAUGCCAAAUACCCAUCUCUGUCAUAUCGAUAACUUAACAGGGAGCGUAAAA------ .....-.......((((.........))))................(((...(((((((...............)))))))..)))...------ ( -9.56, z-score = -0.29, R) >consensus CGAUAGCAGGCUAGCGUAGCCGAAAAUCUGGCGAAUUGUAGCCAGACAUCUGGCUA_GU_CCUUAGUCAAGCCAAUCGCCAGAGCAGGCAGGAGU ........((((....((((((((..((((((........))))))..)))))))).............))))....(((......)))...... (-12.04 = -14.71 + 2.67)
| Location | 19,087,504 – 19,087,597 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 68.89 |
| Shannon entropy | 0.60472 |
| G+C content | 0.49519 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -11.72 |
| Energy contribution | -11.36 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.992514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19087504 93 - 22422827 ACUCUUGCCUGCUCUGGCGAUUGGCUUGACUAAGG-AC-UAGCCAGAUGUCUGGCUACAAUUCGCCAGAUUUCCGGCUACGCUAGCCUGCUAUCG ..........(((((((((((((.(((.....)))-..-(((((((....)))))))))).)))))))).....(((((...))))).))..... ( -33.10, z-score = -2.63, R) >droEre2.scaffold_4690 9348916 93 - 18748788 ACUCCAGCCUGCUCUGACGAUUGGCUUGACUGCGGUAC-UAGCCAAAUGUCUGGCUACAAAUCGCCAG-UUUUCGGCUACGAUAGCCUGCUAUCG .....((((......((((.((((((..((....))..-.)))))).))))((((........)))).-.....)))).((((((....)))))) ( -31.60, z-score = -2.86, R) >droYak2.chrX 17676674 92 - 21770863 AUUUCUGUCUGGUCUGGCGACUGCCUUCACUACG--AC-UAGCCAGAUCUCUGGCUACAAAUCGCCAGAUUUUCGGAAACGAUAGCCUGCUAUCG .((((((...((((((((((......((.....)--).-(((((((....)))))))....))))))))))..))))))((((((....)))))) ( -38.10, z-score = -6.19, R) >droSec1.super_8 1381937 92 - 3762037 ACUCCUGCCUGCUCUGGCGAUUGGUCUGAC-AACG-AC-UAGACAGAUGUCUGGCUAAAAGUCGCCAGAUUUCGGACUACGCUAGCCUGGUAUCG .....((((....((((((..((((((((.-....-.(-(....))..(((((((........))))))).))))))))))))))...))))... ( -30.60, z-score = -2.07, R) >droSim1.chrX 14750149 92 - 17042790 ACUCCUGCCUGCUCUGACGAUUGGCCUGAC-AACG-AC-UAGACAGAUGUCUGGCUAAAAGUAGCCAGAUUUCCGCCUACUCUAGCCUGGUAUCG .....((((.(((..((((.(((......)-))))-..-(((.(.((.(((((((((....))))))))).)).).))).)).)))..))))... ( -25.60, z-score = -1.67, R) >droGri2.scaffold_14853 2231539 88 + 10151454 ------UUUUACGCUCCCUGUUAAGUUAUCGAUAUGACAGAGAUGGGUAUUUGGCAUUGUUUGGUUCGAUAUUUCGCUUCGAUAUUUUG-UAUUA ------...(((.(((.((((((.(((...))).)))))).)).).)))....(((..((((((..(((....)))..)))).))..))-).... ( -16.00, z-score = -0.01, R) >consensus ACUCCUGCCUGCUCUGGCGAUUGGCUUGACUAAGG_AC_UAGACAGAUGUCUGGCUACAAAUCGCCAGAUUUCCGGCUACGAUAGCCUGCUAUCG .................((((.((((.............(((.(.((.(((((((........))))))).)).).)))....))))....)))) (-11.72 = -11.36 + -0.35)


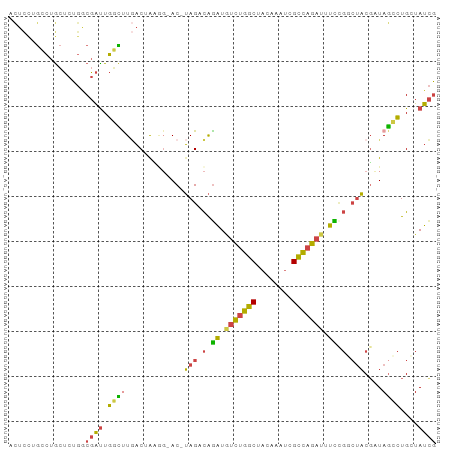
| Location | 19,087,522 – 19,087,624 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 59.12 |
| Shannon entropy | 0.78503 |
| G+C content | 0.44635 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -4.93 |
| Energy contribution | -5.91 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.17 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.766777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
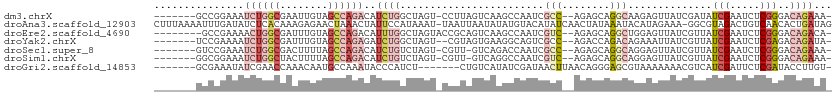
>dm3.chrX 19087522 102 + 22422827 -------GCCGGAAAUCUGGCGAAUUGUAGCCAGACAUCUGGCUAGU-CCUUAGUCAAGCCAAUCGCC--AGAGCAGGCAAGAGUUAUCGAUAUCGAAUCUCGGGACAGAAA- -------((((((..((((((........))))))..))))))..((-(((.......(((..((...--.))...)))..(((...(((....)))..)))))))).....- ( -39.50, z-score = -4.06, R) >droAna3.scaffold_12903 159258 111 - 802071 CUUUAAAAUUUGAUAUCUCACAAAGAGAACUAAACUAUUCCAUAAAU-UAAUUAAUAUAUGUACAUAUCAACUAUAAAUACAUAGAAA-GGCGUAGACUGUCAACACUGAUAG ((((.....((((((((((.....))))............((((.((-(....))).))))....))))))((((......)))))))-).......((((((....)))))) ( -14.40, z-score = -1.15, R) >droEre2.scaffold_4690 9348934 102 + 18748788 --------GCCGAAAACUGGCGAUUUGUAGCCAGACAUUUGGCUAGUACCGCAGUCAAGCCAAUCGUC--AGAGCAGGCUGGAGUUAUCGUUAUCGAAUCUCGGGACAGACA- --------((((((..(((((........)))))...))))))..........(((.((((..((...--.))...)))).(((...(((....)))..)))......))).- ( -30.80, z-score = -1.22, R) >droYak2.chrX 17676692 101 + 21770863 -------UCCGAAAAUCUGGCGAUUUGUAGCCAGAGAUCUGGCUAGU--CGUAGUGAAGGCAGUCGCC--AGACCAGACAGAAAUUAUCGUUAUCGAAUCUCGAGACAGAUA- -------........((((((((((..(((((((....)))))))((--(........))))))))))--)))............((((....(((.....)))....))))- ( -34.50, z-score = -3.38, R) >droSec1.super_8 1381955 101 + 3762037 -------GUCCGAAAUCUGGCGACUUUUAGCCAGACAUCUGUCUAGU-CGUU-GUCAGACCAAUCGCC--AGAGCAGGCAGGAGUUAUCGUUAUCGAAUCUCGGGACAGAAA- -------((((....(((((((((..((((.(((....))).)))).-.)))-))))))......(((--......)))..(((...(((....)))..))).)))).....- ( -34.80, z-score = -2.80, R) >droSim1.chrX 14750167 101 + 17042790 -------GGCGGAAAUCUGGCUACUUUUAGCCAGACAUCUGUCUAGU-CGUU-GUCAGGCCAAUCGUC--AGAGCAGGCAGGAGUUAUCGUUAUCGAAUCUCGGGACAGAAA- -------((((((..((((((((....))))))))..))))))....-..((-(((..(((..((...--.))...)))..(((...(((....)))..)))..)))))...- ( -34.90, z-score = -2.77, R) >droGri2.scaffold_14853 2231556 98 - 10151454 -------GCGAAAUAUCGAACCAAACAAUGCCAAAUACCCAUCU-------CUGUCAUAUCGAUAACUUAACAGGGAGCGUAAAAAAACGUCAUCGAUUCUCGAUACCUUGU- -------((((..((((((......................(((-------((((...............)))))))((((......)))).........))))))..))))- ( -16.06, z-score = -1.02, R) >consensus _______GCCGGAAAUCUGGCGAAUUGUAGCCAGACAUCUGGCUAGU_CCUUAGUCAAGCCAAUCGCC__AGAGCAGGCAGGAGUUAUCGUUAUCGAAUCUCGGGACAGAAA_ ...............((((((........))))))..........................................................(((.....)))......... ( -4.93 = -5.91 + 0.98)
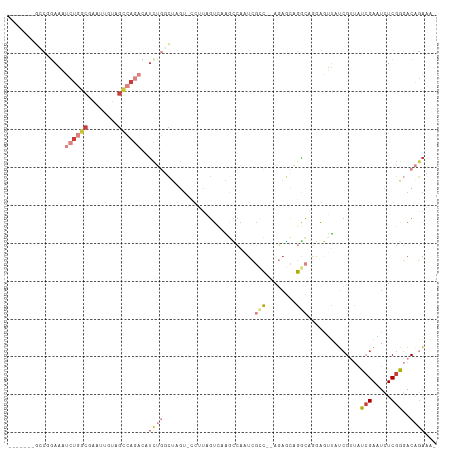
| Location | 19,087,522 – 19,087,624 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 59.12 |
| Shannon entropy | 0.78503 |
| G+C content | 0.44635 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -6.93 |
| Energy contribution | -6.19 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.93 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.667513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
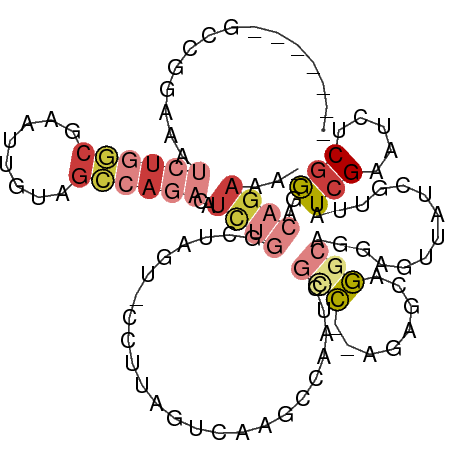
>dm3.chrX 19087522 102 - 22422827 -UUUCUGUCCCGAGAUUCGAUAUCGAUAACUCUUGCCUGCUCU--GGCGAUUGGCUUGACUAAGG-ACUAGCCAGAUGUCUGGCUACAAUUCGCCAGAUUUCCGGC------- -.....((((.(((..(((....)))...)))(((((......--))))).............))-))..(((.((.(((((((........))))))).)).)))------- ( -34.40, z-score = -2.65, R) >droAna3.scaffold_12903 159258 111 + 802071 CUAUCAGUGUUGACAGUCUACGCC-UUUCUAUGUAUUUAUAGUUGAUAUGUACAUAUAUUAAUUA-AUUUAUGGAAUAGUUUAGUUCUCUUUGUGAGAUAUCAAAUUUUAAAG .........((((....(((.((.-.(((((((......(((((((((((....)))))))))))-...)))))))..)).))).((((.....))))..))))......... ( -18.90, z-score = -0.56, R) >droEre2.scaffold_4690 9348934 102 - 18748788 -UGUCUGUCCCGAGAUUCGAUAACGAUAACUCCAGCCUGCUCU--GACGAUUGGCUUGACUGCGGUACUAGCCAAAUGUCUGGCUACAAAUCGCCAGUUUUCGGC-------- -........((((((.(((....)))....((.((((...((.--...))..)))).))(((((((..((((((......))))))...)))).))).)))))).-------- ( -26.50, z-score = -0.48, R) >droYak2.chrX 17676692 101 - 21770863 -UAUCUGUCUCGAGAUUCGAUAACGAUAAUUUCUGUCUGGUCU--GGCGACUGCCUUCACUACG--ACUAGCCAGAUCUCUGGCUACAAAUCGCCAGAUUUUCGGA------- -((((....(((.....)))....))))...((((...(((((--(((((......((.....)--).(((((((....)))))))....))))))))))..))))------- ( -32.50, z-score = -3.16, R) >droSec1.super_8 1381955 101 - 3762037 -UUUCUGUCCCGAGAUUCGAUAACGAUAACUCCUGCCUGCUCU--GGCGAUUGGUCUGAC-AACG-ACUAGACAGAUGUCUGGCUAAAAGUCGCCAGAUUUCGGAC------- -.....((((.(((..(((....)))...)))........(((--(((((((((((.(((-(.(.-........).)))).)))))...)))))))))....))))------- ( -31.60, z-score = -2.32, R) >droSim1.chrX 14750167 101 - 17042790 -UUUCUGUCCCGAGAUUCGAUAACGAUAACUCCUGCCUGCUCU--GACGAUUGGCCUGAC-AACG-ACUAGACAGAUGUCUGGCUAAAAGUAGCCAGAUUUCCGCC------- -..((((((..(((..(((....)))...)))..(((...((.--...))..))).....-....-....)))))).(((((((((....))))))))).......------- ( -27.50, z-score = -2.42, R) >droGri2.scaffold_14853 2231556 98 + 10151454 -ACAAGGUAUCGAGAAUCGAUGACGUUUUUUUACGCUCCCUGUUAAGUUAUCGAUAUGACAG-------AGAUGGGUAUUUGGCAUUGUUUGGUUCGAUAUUUCGC------- -...((((((((((...(((((.((......(((.(((.((((((.(((...))).))))))-------.)).).)))..)).))))).....))))))))))...------- ( -22.10, z-score = -0.26, R) >consensus _UUUCUGUCCCGAGAUUCGAUAACGAUAACUCCUGCCUGCUCU__GGCGAUUGGCUUGACUAAGG_ACUAGACAGAUGUCUGGCUACAAUUCGCCAGAUUUCCGGC_______ ..................................(((...((......))..)))...................((.(((((((........))))))).))........... ( -6.93 = -6.19 + -0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:27 2011