
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,051,775 – 19,051,905 |
| Length | 130 |
| Max. P | 0.874889 |

| Location | 19,051,775 – 19,051,886 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.51 |
| Shannon entropy | 0.38675 |
| G+C content | 0.43045 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -19.27 |
| Energy contribution | -20.83 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.874889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
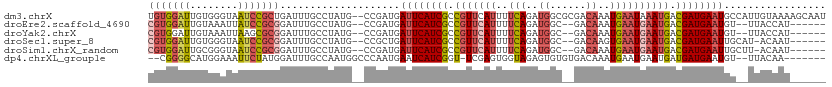
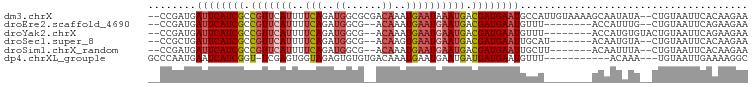
>dm3.chrX 19051775 111 + 22422827 UGUGGAUUGUGGGUAAUCCGCUGAUUUGCCUAUG--CCGAUGAUUCAUCGCCGUUCAUUUUCAGAUGGCGCGACAAAUGAAUAAAUGACGAUGAAUGCCAUUGUAAAAGCAAU .((((((((....))))))))....((((...((--(.((((((((((((.((((....((((..((......))..))))..)))).))))))))..)))))))...)))). ( -29.90, z-score = -0.79, R) >droEre2.scaffold_4690 9315701 101 + 18748788 CGUGGAUUGUAAAUUAUCCGCGGAUUUGCCUAUG--CCGAUGAUUCAUCGCCGUUCAUUUUCAGAUGGC--GACAAAUGAAUGAAUGACGAUGAAUGU--UUACCAU------ .((((((........))))))(((((.((....)--).))).((((((((.(((((((..(((..((..--..))..)))))))))).))))))))..--...))..------ ( -32.20, z-score = -2.78, R) >droYak2.chrX 17643322 101 + 21770863 CGUGGAUUGUAAAUUAAGCGCGGAUUUGCCUAUG--CCGAUGAUUCAUCGCCGUUCAUUUUCAGAUGGC--GACAAAUGAAUGAAUGACGAUGAAUGU--UUACCAU------ .((((...(((((((.......))))))).....--......((((((((.(((((((..(((..((..--..))..)))))))))).))))))))..--...))))------ ( -28.30, z-score = -1.59, R) >droSec1.super_8 1341124 102 + 3762037 CGUGGAUUGUGGGUAAUCCGCGGAUUUGCCUAUG--CCGCUGAUUCAUCGCCGUUCAUUUUCAGAUGGC--GACAAGUGAAUGAAUGACGAUGAAUUGCAU-ACAAU------ .((((...((((((((((....)).)))))))).--)))).(((((((((.(((((((..(((..((..--..))..)))))))))).)))))))))....-.....------ ( -37.60, z-score = -3.10, R) >droSim1.chrX_random 4946916 102 + 5698898 CGUGGAUUGCGGGUAAUCCGCGGAUUUGCCUAUG--CCGAUGAUUCAUCGCCGUUCAUUUUCAGAUGGC--GACAAAUGAAUGAAUGACGAUGAAUUGCUU-ACAAU------ (((((((((....))))))))).(((.((....)--).)))(((((((((.(((((((..(((..((..--..))..)))))))))).)))))))))....-.....------ ( -35.30, z-score = -2.72, R) >dp4.chrXL_group1e 10744896 101 + 12523060 --CGGGGCAUGGAAAUUCUAUGGAUUUGCCAAUGGCCCAAUGAAUCAUCGGU-UCGAGUGGUAGAGUGUGUGACAAAUGAAUGAAUGAUGAUGAAUGU--UUACAA------- --.(((.((((((((((.....))))).)).))).))).((..(((((((((-(((..((.............))..)))))...)))))))..))..--......------- ( -23.92, z-score = -0.33, R) >consensus CGUGGAUUGUGGAUAAUCCGCGGAUUUGCCUAUG__CCGAUGAUUCAUCGCCGUUCAUUUUCAGAUGGC__GACAAAUGAAUGAAUGACGAUGAAUGU__UUACAAU______ (((((((........)))))))....................((((((((.(((((((..(((..((......))..)))))))))).))))))))................. (-19.27 = -20.83 + 1.56)


| Location | 19,051,775 – 19,051,886 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Shannon entropy | 0.38675 |
| G+C content | 0.43045 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -13.74 |
| Energy contribution | -15.55 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
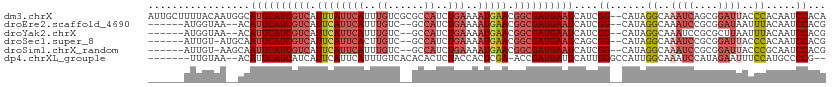
>dm3.chrX 19051775 111 - 22422827 AUUGCUUUUACAAUGGCAUUCAUCGUCAUUUAUUCAUUUGUCGCGCCAUCUGAAAAUGAACGGCGAUGAAUCAUCGG--CAUAGGCAAAUCAGCGGAUUACCCACAAUCCACA .((((((...(.(((..((((((((((.((((((((..((......))..)))..))))).))))))))))))).).--...))))))....(.(((((......))))).). ( -28.40, z-score = -1.71, R) >droEre2.scaffold_4690 9315701 101 - 18748788 ------AUGGUAA--ACAUUCAUCGUCAUUCAUUCAUUUGUC--GCCAUCUGAAAAUGAACGGCGAUGAAUCAUCGG--CAUAGGCAAAUCCGCGGAUAAUUUACAAUCCACG ------...((((--(.((((((((((.((((((((..((..--..))..)))..))))).)))))))))).(((.(--(...((.....)))).)))..)))))........ ( -25.00, z-score = -1.65, R) >droYak2.chrX 17643322 101 - 21770863 ------AUGGUAA--ACAUUCAUCGUCAUUCAUUCAUUUGUC--GCCAUCUGAAAAUGAACGGCGAUGAAUCAUCGG--CAUAGGCAAAUCCGCGCUUAAUUUACAAUCCACG ------...((((--(.((((((((((.((((((((..((..--..))..)))..))))).))))))))))......--..(((((........))))).)))))........ ( -25.10, z-score = -2.14, R) >droSec1.super_8 1341124 102 - 3762037 ------AUUGU-AUGCAAUUCAUCGUCAUUCAUUCACUUGUC--GCCAUCUGAAAAUGAACGGCGAUGAAUCAGCGG--CAUAGGCAAAUCCGCGGAUUACCCACAAUCCACG ------.((((-((((.((((((((((.((((((((..((..--..))..)))..))))).)))))))))).....)--)))..))))...((.(((((......))))).)) ( -29.60, z-score = -2.53, R) >droSim1.chrX_random 4946916 102 - 5698898 ------AUUGU-AAGCAAUUCAUCGUCAUUCAUUCAUUUGUC--GCCAUCUGAAAAUGAACGGCGAUGAAUCAUCGG--CAUAGGCAAAUCCGCGGAUUACCCGCAAUCCACG ------.((((-..((.((((((((((.((((((((..((..--..))..)))..))))).)))))))))).....)--)....))))....((((.....))))........ ( -28.90, z-score = -2.21, R) >dp4.chrXL_group1e 10744896 101 - 12523060 -------UUGUAA--ACAUUCAUCAUCAUUCAUUCAUUUGUCACACACUCUACCACUCGA-ACCGAUGAUUCAUUGGGCCAUUGGCAAAUCCAUAGAAUUUCCAUGCCCCG-- -------......--......((((((.(((...........................))-)..)))))).....((((...(((.((.((....)).)).))).))))..-- ( -13.13, z-score = 0.42, R) >consensus ______AUUGUAA__ACAUUCAUCGUCAUUCAUUCAUUUGUC__GCCAUCUGAAAAUGAACGGCGAUGAAUCAUCGG__CAUAGGCAAAUCCGCGGAUUACCCACAAUCCACG .................((((((((((.((((((((..((......))..)))..))))).))))))))))....((......((..((((....))))..)).....))... (-13.74 = -15.55 + 1.81)
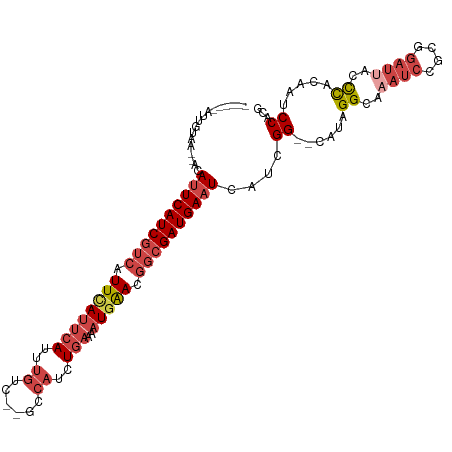
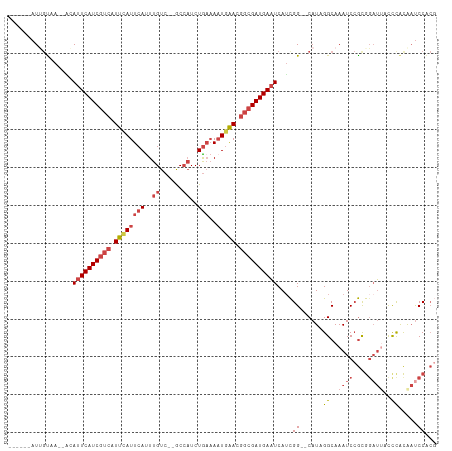
| Location | 19,051,809 – 19,051,905 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 78.69 |
| Shannon entropy | 0.37540 |
| G+C content | 0.38195 |
| Mean single sequence MFE | -21.47 |
| Consensus MFE | -14.15 |
| Energy contribution | -15.23 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.829130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19051809 96 + 22422827 --CCGAUGAUUCAUCGCCGUUCAUUUUCAGAUGGCGCGACAAAUGAAUAAAUGACGAUGAAUGCCAUUGUAAAAGCAAUAUA--CUGUAAUUCACAAGAA --.(((((((((((((.((((....((((..((......))..))))..)))).))))))))..))))).....(((.....--.)))............ ( -18.70, z-score = -0.36, R) >droEre2.scaffold_4690 9315735 86 + 18748788 --CCGAUGAUUCAUCGCCGUUCAUUUUCAGAUGGCG--ACAAAUGAAUGAAUGACGAUGAAUGUUU--------ACCAUUUG--CUGUAAUUCAGAAGAA --......((((((((.(((((((..(((..((...--.))..)))))))))).))))))))....--------....(((.--(((.....))).))). ( -23.00, z-score = -2.07, R) >droYak2.chrX 17643356 88 + 21770863 --CCGAUGAUUCAUCGCCGUUCAUUUUCAGAUGGCG--ACAAAUGAAUGAAUGACGAUGAAUGUUU--------ACCAUGUGUACUGUAAUUCAGAAGAA --......((((((((.(((((((..(((..((...--.))..)))))))))).))))))))....--------..........(((.....)))..... ( -23.20, z-score = -1.92, R) >droSec1.super_8 1341158 87 + 3762037 --CCGCUGAUUCAUCGCCGUUCAUUUUCAGAUGGCG--ACAAGUGAAUGAAUGACGAUGAAUUGCAU-------ACAAUGUA--CUGUAAUUCACAAGAA --..((.(((((((((.(((((((..(((..((...--.))..)))))))))).))))))))))).(-------(((.....--.))))........... ( -25.90, z-score = -2.74, R) >droSim1.chrX_random 4946950 87 + 5698898 --CCGAUGAUUCAUCGCCGUUCAUUUUCAGAUGGCG--ACAAAUGAAUGAAUGACGAUGAAUUGCUU-------ACAAUUUA--CUGUAAUUCACAAGAA --..((.(((((((((.(((((((..(((..((...--.))..)))))))))).)))))))))..((-------(((.....--.))))).))....... ( -24.50, z-score = -3.26, R) >dp4.chrXL_group1e 10744928 85 + 12523060 GCCCAAUGAAUCAUCGGU-UCGAGUGGUAGAGUGUGUGACAAAUGAAUGAAUGAUGAUGAAUGUUU-----------ACAAA---UGUAAUUGAAAAGGC (((..((..(((((((((-(((..((.............))..)))))...)))))))..))..((-----------((...---.)))).......))) ( -13.52, z-score = 0.18, R) >consensus __CCGAUGAUUCAUCGCCGUUCAUUUUCAGAUGGCG__ACAAAUGAAUGAAUGACGAUGAAUGUCU________ACAAUAUA__CUGUAAUUCACAAGAA ........((((((((.(((((((..(((..((......))..)))))))))).))))))))...................................... (-14.15 = -15.23 + 1.08)

| Location | 19,051,809 – 19,051,905 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 78.69 |
| Shannon entropy | 0.37540 |
| G+C content | 0.38195 |
| Mean single sequence MFE | -19.52 |
| Consensus MFE | -12.95 |
| Energy contribution | -14.37 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19051809 96 - 22422827 UUCUUGUGAAUUACAG--UAUAUUGCUUUUACAAUGGCAUUCAUCGUCAUUUAUUCAUUUGUCGCGCCAUCUGAAAAUGAACGGCGAUGAAUCAUCGG-- ..((.((((.......--.....((((........))))(((((((((.((((((((..((......))..)))..))))).))))))))))))).))-- ( -21.40, z-score = -1.02, R) >droEre2.scaffold_4690 9315735 86 - 18748788 UUCUUCUGAAUUACAG--CAAAUGGU--------AAACAUUCAUCGUCAUUCAUUCAUUUGU--CGCCAUCUGAAAAUGAACGGCGAUGAAUCAUCGG-- .....((((.((((..--......))--------))..((((((((((.((((((((..((.--...))..)))..))))).))))))))))..))))-- ( -21.30, z-score = -2.36, R) >droYak2.chrX 17643356 88 - 21770863 UUCUUCUGAAUUACAGUACACAUGGU--------AAACAUUCAUCGUCAUUCAUUCAUUUGU--CGCCAUCUGAAAAUGAACGGCGAUGAAUCAUCGG-- .....((((.((((.((....)).))--------))..((((((((((.((((((((..((.--...))..)))..))))).))))))))))..))))-- ( -20.50, z-score = -1.98, R) >droSec1.super_8 1341158 87 - 3762037 UUCUUGUGAAUUACAG--UACAUUGU-------AUGCAAUUCAUCGUCAUUCAUUCACUUGU--CGCCAUCUGAAAAUGAACGGCGAUGAAUCAGCGG-- ...........(((((--....))))-------)(((.((((((((((.((((((((..((.--...))..)))..))))).))))))))))..))).-- ( -22.30, z-score = -1.72, R) >droSim1.chrX_random 4946950 87 - 5698898 UUCUUGUGAAUUACAG--UAAAUUGU-------AAGCAAUUCAUCGUCAUUCAUUCAUUUGU--CGCCAUCUGAAAAUGAACGGCGAUGAAUCAUCGG-- ..((.((((.((((((--....))))-------))....(((((((((.((((((((..((.--...))..)))..))))).))))))))))))).))-- ( -22.70, z-score = -2.52, R) >dp4.chrXL_group1e 10744928 85 - 12523060 GCCUUUUCAAUUACA---UUUGU-----------AAACAUUCAUCAUCAUUCAUUCAUUUGUCACACACUCUACCACUCGA-ACCGAUGAUUCAUUGGGC ((((......((((.---...))-----------))......((((((.(((...........................))-)..)))))).....)))) ( -8.93, z-score = -0.43, R) >consensus UUCUUGUGAAUUACAG__UAAAUUGU________AAACAUUCAUCGUCAUUCAUUCAUUUGU__CGCCAUCUGAAAAUGAACGGCGAUGAAUCAUCGG__ ......................................((((((((((.((((((((..((......))..)))..))))).))))))))))........ (-12.95 = -14.37 + 1.42)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:22 2011