
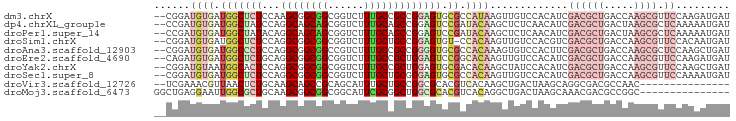
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,045,537 – 19,045,631 |
| Length | 94 |
| Max. P | 0.555344 |

| Location | 19,045,537 – 19,045,631 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 72.53 |
| Shannon entropy | 0.58748 |
| G+C content | 0.59592 |
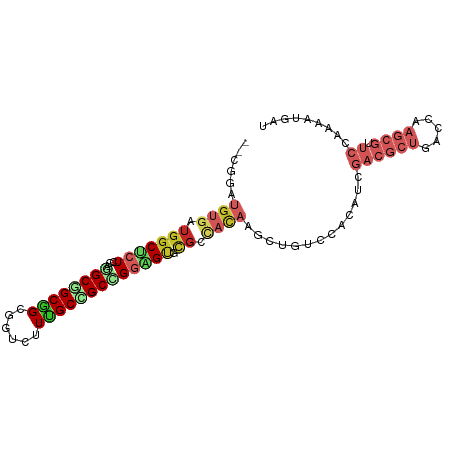
| Mean single sequence MFE | -34.25 |
| Consensus MFE | -15.65 |
| Energy contribution | -17.35 |
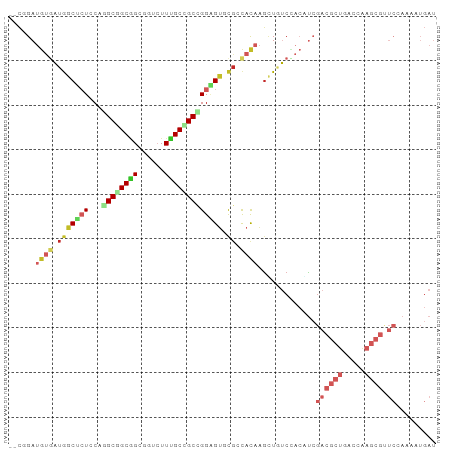
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.555344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19045537 94 - 22422827 --CGGAUGUGAUGGCUCUCCAAGCGGCGGCGGUCUUUGCCGCCGGAGUGCGCCAUAAGUUGUCCACAUCGACGCUGACCAAGCGUUCCAAGAUGAU --.(((((..(((((.((((....(((((((.....)))))))))))...)))))....))))).((((((((((.....))))))....)))).. ( -40.20, z-score = -2.41, R) >dp4.chrXL_group1e 10738893 94 - 12523060 --CCGAUGUGAUGGCUAUCCAGGCAGCAGCGGUCUUUGCAGCCGGAGUCCGAUACAAGCUCUCAACAUCGACGCUGACUAAGCGCUCAAAAAUGAU --..(((((((.((((.(((.(((.((((......)))).))))))((.....)).)))).)).)))))((((((.....)))).))......... ( -28.70, z-score = -0.77, R) >droPer1.super_14 172805 94 - 2168203 --CCGAUGUGAUGGCUAUACAGGCAGCAGCGGUCUUUGCAGCCGGAGUCCGAUACAAGCUCUCAACAUCGACGCUGACUAAGCGCUCAAAAAUGAU --..(((((((.((((.....(((.((((......)))).)))((...))......)))).)).)))))((((((.....)))).))......... ( -26.80, z-score = -0.39, R) >droSim1.chrX 14714529 93 - 17042790 --CGGAUGUGAUGGCUCUCCAGGCGGCGGCGGUCUUUGCUGCCGGAGUGU-CCACAAGUUGUCCACGUCGACGCUGACCAAGCGUUCCACAAUGAU --..(((((((..(((((((.((((((((......))))))))))))((.-...)))))..).))))))((((((.....)))))).......... ( -35.60, z-score = -0.83, R) >droAna3.scaffold_12903 111765 94 + 802071 --CGGAUGUGAUGGCCCUCCAGGCGGCGGCCGUCUUUGCCGCCGGGGUGCGCCACAAAGUGUCCACUUCGACGCUGACCAAGCGCUCCAAGCUGAU --.(((..(..(((((((((.((((((((......)))))))))))).).))))....)..))).(((.((((((.....)))).)).)))..... ( -39.60, z-score = -0.85, R) >droEre2.scaffold_4690 9309853 94 - 18748788 --CAGAUGUGAUGGCUCUGCAGGCGGCGGCGGUCUUUGCCGCUGGAGUCCGGCACAAGUUGUCCACAUCGACGCUGACCAAGCGUUCCAAGAUGAU --..(((((((..(((.(((.(((..(((((((....)))))))..).)).)))..)))..).))))))((((((.....)))))).......... ( -38.40, z-score = -1.51, R) >droYak2.chrX 17637439 94 - 21770863 --CGGAUGUAAUGGCACUCCAGGCGGCGGCGGUCUUUGCCGCUGGAGUGCGACACAAGCUAUCCACAUCGACGCUGACCAAGCGUUCCAAGCUGAU --.((((((..(.(((((((((.((((((......))))))))))))))).).....)).)))).....((((((.....)))))).......... ( -37.70, z-score = -1.92, R) >droSec1.super_8 1334907 94 - 3762037 --CGGAUGUGAUGGCUCUCCAGGCGGCGGCGGUCUUUGCUGCGGGAGUGCGCCACAAGUUGUCCACAUCGACGCUGACCAAGCGUUCCAAAAUGAU --..((((((..(((.((((..(((((((......))))))).))))...)))((.....)).))))))((((((.....)))))).......... ( -34.20, z-score = -0.30, R) >droVir3.scaffold_12726 975506 79 - 2840439 --UCGAAACGUUAACUCUGCAAGCAGCCGCAGCAUUUGCUGCCGGCGCACGUCACAAGCUGACUAAGCAGGCGACGCCAAC--------------- --......((((...(((((..((.((((((((....)))).))))))..((((.....))))...))))).)))).....--------------- ( -28.30, z-score = -1.59, R) >droMoj3.scaffold_6473 7752654 81 + 16943266 GGCUGAGGAAUUGGCGCUGCAAGCGGCGGCGGCAUUCGCGGCUGGCGCACGUCACAGGCUGACUAAGCAAACGACGCCGGC--------------- (((((..((((.(.((((((.....)))))).))))).)))))((((..(((.....(((.....)))..))).))))...--------------- ( -33.00, z-score = -0.26, R) >consensus __CGGAUGUGAUGGCUCUCCAGGCGGCGGCGGUCUUUGCCGCCGGAGUGCGCCACAAGCUGUCCACAUCGACGCUGACCAAGCGUUCCAAAAUGAU ......((((.(((((((...((((((((......))))))))))))).)).))))...............((((.....))))............ (-15.65 = -17.35 + 1.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:19 2011