
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,030,874 – 19,030,950 |
| Length | 76 |
| Max. P | 0.995872 |

| Location | 19,030,874 – 19,030,950 |
|---|---|
| Length | 76 |
| Sequences | 6 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 60.67 |
| Shannon entropy | 0.72849 |
| G+C content | 0.53589 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -16.07 |
| Energy contribution | -16.77 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.995872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
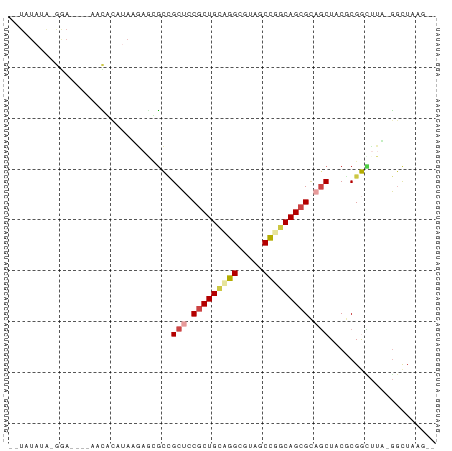
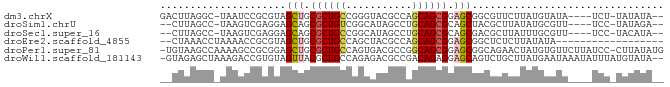
>dm3.chrX 19030874 76 + 22422827 --UAUAUA-AGA----UAUACAUAAGAACGCCGCUCCGCUGCUGGCGUACCCGGCAGCGCAGCUACGCGGAUUA-GCCUAAGUC --......-.((----(.((..(((...(((.(((.(((((((((.....))))))))).)))...)))..)))-...)).))) ( -25.20, z-score = -1.23, R) >droSim1.chrU 2070196 74 + 15797150 --UCUAUA-GGA----AACGCAUAUAAGCGUAGCUGCGCUGCAGGCUAUGCCGACAGCGCUGCUCCUCGACUUA-GGCUAAG-- --......-(..----..)((...((((((.(((.((((((..(((...)))..)))))).)))...)).))))-.))....-- ( -28.90, z-score = -2.02, R) >droSec1.super_16 19905 74 + 1878335 --UAUGUA-GGA----AACGCAAAUAAGCGUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUCGACUUA-GGCUAAG-- --..((.(-(((----.((((......)))).((.(((((((.(((...))).))))))).)))))))).....-.......-- ( -30.10, z-score = -1.59, R) >droEre2.scaffold_4855 234980 64 - 242605 ------------------UAUAUAAGAGAGCCGCUCCGCUGCUGGCGUAGCUGGCAGCGCAGCUACGCGGUUUUAGGUUUAG-- ------------------........((((((((..((((((..((...))..)))))).......))))))))........-- ( -25.10, z-score = -0.97, R) >droPer1.super_81 10290 82 + 277874 CAUAUAAG-GGAUAAGAACACAUAGUUCUGCCGCUCCGCUGCCGGCGUCACUGGCAGCGCAGCUCCGCGGCUUUUGGCUUACA- ........-((...(((((.....))))).))(((.(((((((((.....))))))))).))).....(((.....)))....- ( -30.50, z-score = -1.50, R) >droWil1.scaffold_181143 551640 81 + 1323777 --UAUACAUAAAUAUUUAUUCAUAAGCAGACUGCUCCUCUGUCGGCGUCUCUGGCAGCGUAACUACACGGUCUUUAGCUCUAC- --......................(((((((((.....(((((((.....))))))).((....)).))))))...)))....- ( -14.40, z-score = -0.30, R) >consensus __UAUAUA_GGA____AACACAUAAGAGCGCCGCUCCGCUGCAGGCGUAGCCGGCAGCGCAGCUACGCGGCUUA_GGCUAAG__ ................................(((.(((((((((.....))))))))).)))..................... (-16.07 = -16.77 + 0.70)


| Location | 19,030,874 – 19,030,950 |
|---|---|
| Length | 76 |
| Sequences | 6 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 60.67 |
| Shannon entropy | 0.72849 |
| G+C content | 0.53589 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -11.81 |
| Energy contribution | -12.23 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.903847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
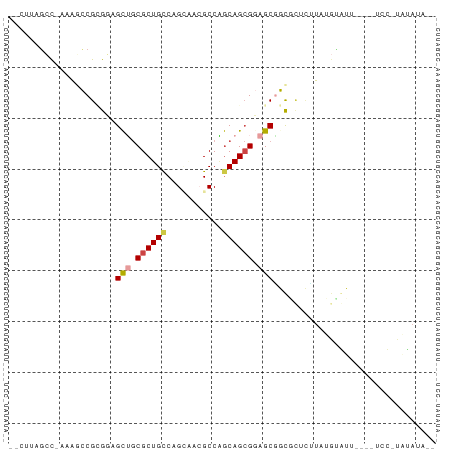
>dm3.chrX 19030874 76 - 22422827 GACUUAGGC-UAAUCCGCGUAGCUGCGCUGCCGGGUACGCCAGCAGCGGAGCGGCGUUCUUAUGUAUA----UCU-UAUAUA-- ......((.-....))((((.(((.((((((.((.....)).)))))).))).))))...((((((..----...-))))))-- ( -26.20, z-score = -0.29, R) >droSim1.chrU 2070196 74 - 15797150 --CUUAGCC-UAAGUCGAGGAGCAGCGCUGUCGGCAUAGCCUGCAGCGCAGCUACGCUUAUAUGCGUU----UCC-UAUAGA-- --.......-....((.((((((.(((((((.(((...))).))))))).)).((((......)))).----)))-)...))-- ( -28.50, z-score = -1.51, R) >droSec1.super_16 19905 74 - 1878335 --CUUAGCC-UAAGUCGAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGACGCUUAUUUGCGUU----UCC-UACAUA-- --.......-.......((((((.(((((((.(((...))).))))))).))(((((......)))))----)))-).....-- ( -31.70, z-score = -2.62, R) >droEre2.scaffold_4855 234980 64 + 242605 --CUAAACCUAAAACCGCGUAGCUGCGCUGCCAGCUACGCCAGCAGCGGAGCGGCUCUCUUAUAUA------------------ --..............((...(((.((((((..((...))..)))))).))).))...........------------------ ( -19.80, z-score = -0.43, R) >droPer1.super_81 10290 82 - 277874 -UGUAAGCCAAAAGCCGCGGAGCUGCGCUGCCAGUGACGCCGGCAGCGGAGCGGCAGAACUAUGUGUUCUUAUCC-CUUAUAUG -((((((......(((.....(((.(((((((.(.....).))))))).))))))(((((.....))))).....-)))))).. ( -30.80, z-score = -1.46, R) >droWil1.scaffold_181143 551640 81 - 1323777 -GUAGAGCUAAAGACCGUGUAGUUACGCUGCCAGAGACGCCGACAGAGGAGCAGUCUGCUUAUGAAUAAAUAUUUAUGUAUA-- -...((((........(.((((.....)))))..((((.((......))....))))))))((((((....)))))).....-- ( -14.60, z-score = 0.57, R) >consensus __CUUAGCC_AAAGCCGCGGAGCUGCGCUGCCAGCAACGCCAGCAGCGGAGCGGCGCUCUUAUGUAUU____UCC_UAUAUA__ .....................(((.((((((...........)))))).)))................................ (-11.81 = -12.23 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:18 2011