
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,028,894 – 19,028,984 |
| Length | 90 |
| Max. P | 0.796313 |

| Location | 19,028,894 – 19,028,984 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 69.28 |
| Shannon entropy | 0.58042 |
| G+C content | 0.31881 |
| Mean single sequence MFE | -15.41 |
| Consensus MFE | -7.05 |
| Energy contribution | -6.53 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.796313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
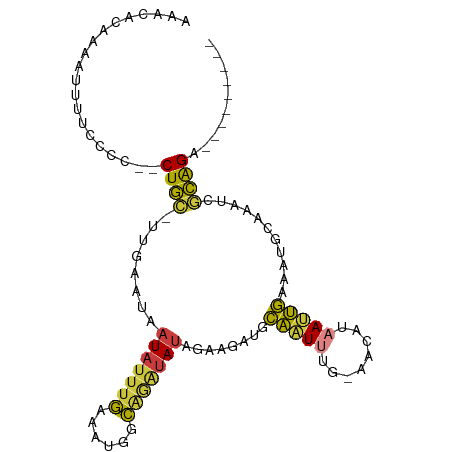
>dm3.chrX 19028894 90 - 22422827 AAACACAAAAUUUUCCCC--CUGC-UUGAAUAAUAUUUGAAAUGGCAGAUAUGGAAGAUGCAAUUUG-AACAUAAUUGAAAUGCAAAUCGCAGA--------- ..................--((((-.......(((((((......)))))))......((((.((..-(......)..)).))))....)))).--------- ( -15.00, z-score = -0.76, R) >droSim1.chrX 14702281 90 - 17042790 AAACACAAAAUGUACCCC--CUGC-UUGAAUAAUAUUUGAAAUGGCAGAUAUAGAAGAUGCAAUUUG-AACAUAAUUGAAAUGCAAAUCGCAGA--------- ..................--((((-.......(((((((......)))))))......((((.((..-(......)..)).))))....)))).--------- ( -15.00, z-score = -1.64, R) >droSec1.super_8 1325231 90 - 3762037 AAACACAAAAUUUACCCU--CUGC-UUGAAUAAUAUUUGAAAUGGCAGAUAUAGAAGAUGCAAUUUG-AACAUAAUUGAAAUGCAAAUCGCAGA--------- .................(--((((-.......(((((((......)))))))......((((.((..-(......)..)).))))....)))))--------- ( -15.60, z-score = -1.61, R) >droYak2.chrX 17627214 92 - 21770863 AAACACAAAAUUUUCCCCCUCUGC-CUGAAUAAUAUUUGAAAUGGCAGAUAUAGAUGAUGCAAUUUG-AACAUAAUUGAAAUGCAAAUCGCAGA--------- ...................(((((-.......(((((((......))))))).(((..((((.((..-(......)..)).)))).))))))))--------- ( -17.50, z-score = -2.06, R) >droEre2.scaffold_4690 9300529 92 - 18748788 AAACACAAAAAGUUCCCCCACUGC-UUGAAUAAUAUUUGAAAUGGCAGAUAUAGAAGAUGCAAUUUG-AACAUAAUUGAAAUGCAAAUCGCAGA--------- ....................((((-.......(((((((......)))))))......((((.((..-(......)..)).))))....)))).--------- ( -15.20, z-score = -1.34, R) >droAna3.scaffold_12903 102029 102 + 802071 AAUUACUAGAUUUUUAGCUGCUUUGUUUAGUUAUAUUUG-AGUACCGAGUAUAAAUCAUAAAAUGAACAUCUUAACUUAAAUGCAAAAAAGGGAAAUUGCAGA .................((((..((((((.(((((((((-.....))))))))).........))))))..((..(((..........)))..))...)))). ( -13.00, z-score = 0.81, R) >droWil1.scaffold_181096 8221419 81 - 12416693 ----CCUAAAAUGUUCUCUGCUG--UUUGCAACUAAUCAAAGUAAUA---AUAACUAAUUCGGU----UGCCAAACCGCAUCGCAAAUAACAGG--------- ----.............(((.((--(((((.(((......)))....---..........((((----(....)))))....))))))).))).--------- ( -16.60, z-score = -3.20, R) >consensus AAACACAAAAUUUUCCCC__CUGC_UUGAAUAAUAUUUGAAAUGGCAGAUAUAGAAGAUGCAAUUUG_AACAUAAUUGAAAUGCAAAUCGCAGA_________ ....................((((........(((((((......)))))))........(((((........)))))...........)))).......... ( -7.05 = -6.53 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:17 2011