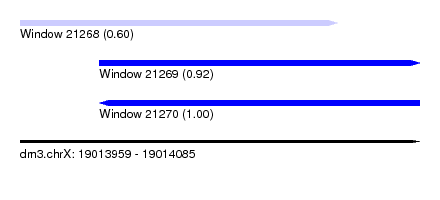
| Sequence ID | dm3.chrX |
|---|---|
| Location | 19,013,959 – 19,014,085 |
| Length | 126 |
| Max. P | 0.996865 |

| Location | 19,013,959 – 19,014,059 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 65.82 |
| Shannon entropy | 0.68347 |
| G+C content | 0.56129 |
| Mean single sequence MFE | -30.49 |
| Consensus MFE | -10.55 |
| Energy contribution | -10.97 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
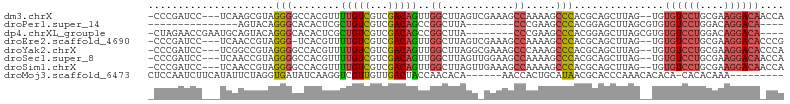
>dm3.chrX 19013959 100 + 22422827 -CCCGAUCC---UCAAGCGUAGGGGCCACGUUUUGUCGUCGACAGUUGGCUUAGUCGAAAGCCAAAAGCCCACGCAGCUUAG--UGUGUCCUGCGAAGGACAACCA -...(.(((---(....(((((((((......(((((...)))))(((((((......)))))))..)))(((((......)--)))).)))))).)))))..... ( -32.70, z-score = -1.40, R) >droPer1.super_14 2012804 79 - 2168203 ---------------AGUACAGGGCACACUCGCUGUCGUCGACAGCCGGCUUA--------CCCGAAGCCCACGGAGCUUAGCGUGUGUCCUGGACAGGACA---- ---------------.((.((((((((((.(((((((...)))))).(((((.--------....)))))...........).)))))))))).))......---- ( -33.80, z-score = -2.09, R) >dp4.chrXL_group1e 1801401 93 - 12523060 -CUAGAACCGAAUGCAGUACAGGGCACACUCGCUGUCGUCGACAGCCGGCUUA--------CCCGAAGCCCACGGAGCUUAGCGUGUGUCCUGGACAGGACA---- -...............((.((((((((((.(((((((...)))))).(((((.--------....)))))...........).)))))))))).))......---- ( -33.80, z-score = -1.20, R) >droEre2.scaffold_4690 9285765 99 + 18748788 -CCCGAUCC---UCAACCGUAGGG-UCACGUUUUGUCGUCGACAGUUGGCUUAGUCGAAAGCCAAAAGCCCACGCAGCUUAG--UGUGUCCUGCGAAGGACACCCG -...(((((---((....).))))-)).....(((.(((.(.(..(((((((......)))))))..).).))))))....(--.(((((((....))))))).). ( -31.30, z-score = -1.77, R) >droYak2.chrX 17611371 100 + 21770863 -CCCGAUCC---UCGGCCGUAGGGGCCACGUUUUGUCGUCGACAGUUGGCUUAGGCGAAAGCCCAAAGCCCACGCAGCUUAG--UGUGUCCUGCGAAGGACACCCA -........---..((((.....))))......((((.(((.(((..(((((.(((....)))..)))))(((((......)--))))..))))))..)))).... ( -37.30, z-score = -1.40, R) >droSec1.super_8 1304377 100 + 3762037 -CCCGAUCC---UCAACCGUAGGGGCCACGUUUUGUCGUCGACAGUUGGCUUAGUUGGAAGCCAAAAGCCCACGCAGCUUAG--UGUGUCCUGCGAAGGACAACCA -...(.(((---(....(((((((((......(((((...)))))(((((((......)))))))..)))(((((......)--)))).)))))).)))))..... ( -31.70, z-score = -0.79, R) >droSim1.chrX 14691897 100 + 17042790 -CCCGAUCC---UCAACCGUAGGGGCCACGUUUUGUCGUCGACAGUUGGCUUAGUUGAAAGCCAAAAGCCCACGCAGCUUAG--UGUGUCCUGCGAAGGACAACCA -...(.(((---(....(((((((((......(((((...)))))(((((((......)))))))..)))(((((......)--)))).)))))).)))))..... ( -31.60, z-score = -1.24, R) >droMoj3.scaffold_6473 7706832 90 - 16943266 CUCCAAUCUUCAUAUUCUAGGUGAUAUCAAGGUCGUUGUUGACUACCAACACA------AACCACUGCAUAACGCACCCAAACACACA-CACACAAA--------- ...................((((.(((((.(((...(((((.....)))))..------.)))..)).)))...))))..........-........--------- ( -11.70, z-score = -0.66, R) >consensus _CCCGAUCC___UCAACCGUAGGGGCCACGUUUUGUCGUCGACAGUUGGCUUAGU_GAAAGCCAAAAGCCCACGCAGCUUAG__UGUGUCCUGCGAAGGACA_CCA .....................(((........(((((...)))))..((............)).....)))...............((((((....)))))).... (-10.55 = -10.97 + 0.42)
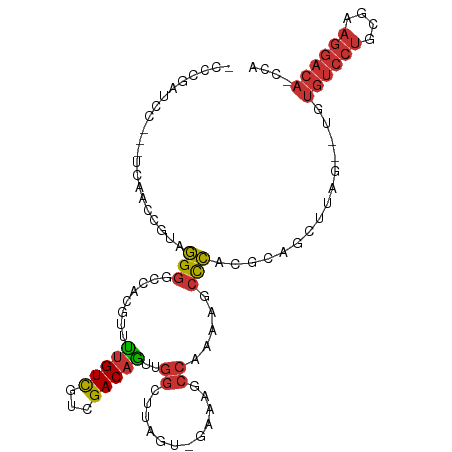
| Location | 19,013,984 – 19,014,085 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 64.85 |
| Shannon entropy | 0.68737 |
| G+C content | 0.55047 |
| Mean single sequence MFE | -33.96 |
| Consensus MFE | -12.04 |
| Energy contribution | -12.58 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19013984 101 + 22422827 -GUUUUGUCGUCGACAGUUGGCUUAGUCGAAAGCCAAAAGCCCACGCAGCUUAG--UGUGUCCUGCGAAGGACAACCACCCAAGGGGGUGGUGGAGCUGAAAGG -...(((((...)))))(((((((......)))))))....((...((((((..--..((((((....))))))(((((((....))))))).))))))...)) ( -41.00, z-score = -2.79, R) >droPer1.super_14 2012818 91 - 2168203 -UCGCUGUCGUCGACAGCCGGCUUA--------CCCGAAGCCCACGGAGCUUAGCGUGUGUCCUGGACAGGACACCCAAGGGAGAAGUUCAACAACGAGU---- -..((((((...)))))).(((((.--------....)))))....((((((..(.((((((((....)))))))....).)..))))))..........---- ( -30.70, z-score = -0.84, R) >dp4.chrXL_group1e 1801429 91 - 12523060 -UCGCUGUCGUCGACAGCCGGCUUA--------CCCGAAGCCCACGGAGCUUAGCGUGUGUCCUGGACAGGACACCCAAGGGAGAAGUUCAACAACGAGA---- -..((((((...)))))).(((((.--------....)))))....((((((..(.((((((((....)))))))....).)..))))))..........---- ( -30.70, z-score = -0.81, R) >droEre2.scaffold_4690 9285789 101 + 18748788 -GUUUUGUCGUCGACAGUUGGCUUAGUCGAAAGCCAAAAGCCCACGCAGCUUAG--UGUGUCCUGCGAAGGACACCCGCCCAAUGGGUUGGGUGAGCUGAAAGG -...(((((...)))))(((((((......)))))))....((...((((((.(--.(((((((....))))))).)((((((....))))))))))))...)) ( -38.80, z-score = -2.40, R) >droYak2.chrX 17611396 93 + 21770863 -GUUUUGUCGUCGACAGUUGGCUUAGGCGAAAGCCCAAAGCCCACGCAGCUUAG--UGUGUCCUGCGAAGGACACCCACCCAAUGGGUCUGAAAGU-------- -.....(((.(((.(((..(((((.(((....)))..)))))(((((......)--))))..))))))..)))(((((.....)))))........-------- ( -34.00, z-score = -2.54, R) >droSec1.super_8 1304402 101 + 3762037 -GUUUUGUCGUCGACAGUUGGCUUAGUUGGAAGCCAAAAGCCCACGCAGCUUAG--UGUGUCCUGCGAAGGACAACCACCCAGGGGGGUGGUGAAGCUGAAAGG -...(((((...)))))(((((((......)))))))....((...((((((..--..((((((....))))))(((((((....))))))).))))))...)) ( -40.70, z-score = -2.42, R) >droSim1.chrX 14691922 101 + 17042790 -GUUUUGUCGUCGACAGUUGGCUUAGUUGAAAGCCAAAAGCCCACGCAGCUUAG--UGUGUCCUGCGAAGGACAACCACCCAGGAGGGUGGUGGAGCUGAAAGG -...(((((...)))))(((((((......)))))))....((...((((((..--..((((((....))))))(((((((....))))))).))))))...)) ( -41.10, z-score = -3.04, R) >droMoj3.scaffold_6473 7706860 89 - 16943266 AAGGUCGUUGUUGACUACCAACACA------AACCACUGCAUAACGCACCCAAA--CACACACACACAAACAUAGAGAGCAAGGGGAGUUGUAAGAC------- ..(((...(((((.....)))))..------.))).((..(((((...(((...--..........................)))..))))).))..------- ( -14.65, z-score = 0.20, R) >consensus _GUUUUGUCGUCGACAGUUGGCUUAGU_GAAAGCCAAAAGCCCACGCAGCUUAG__UGUGUCCUGCGAAGGACACCCACCCAAGGGGGUGGUGAAGCUGA____ ....(((((...)))))..(((((.............)))))....((((((......((((((....))))))..........)))))).............. (-12.04 = -12.58 + 0.55)
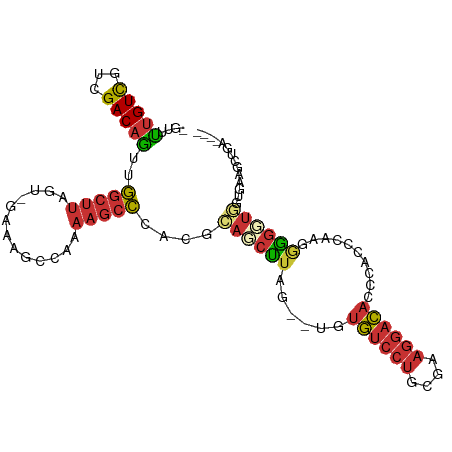
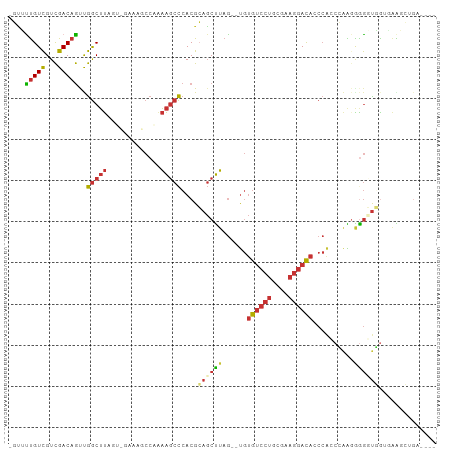
| Location | 19,013,984 – 19,014,085 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 64.85 |
| Shannon entropy | 0.68737 |
| G+C content | 0.55047 |
| Mean single sequence MFE | -35.51 |
| Consensus MFE | -13.55 |
| Energy contribution | -13.17 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.996865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 19013984 101 - 22422827 CCUUUCAGCUCCACCACCCCCUUGGGUGGUUGUCCUUCGCAGGACACA--CUAAGCUGCGUGGGCUUUUGGCUUUCGACUAAGCCAACUGUCGACGACAAAAC- .....(((((..(((((((....)))))))((((((....))))))..--...)))))(((.(((..(((((((......)))))))..))).))).......- ( -41.90, z-score = -4.52, R) >droPer1.super_14 2012818 91 + 2168203 ----ACUCGUUGUUGAACUUCUCCCUUGGGUGUCCUGUCCAGGACACACGCUAAGCUCCGUGGGCUUCGGG--------UAAGCCGGCUGUCGACGACAGCGA- ----..(((((((((.........((((((((((((....)))))))...)))))....((.(((((....--------.))))).))......)))))))))- ( -34.10, z-score = -0.93, R) >dp4.chrXL_group1e 1801429 91 + 12523060 ----UCUCGUUGUUGAACUUCUCCCUUGGGUGUCCUGUCCAGGACACACGCUAAGCUCCGUGGGCUUCGGG--------UAAGCCGGCUGUCGACGACAGCGA- ----..(((((((((.........((((((((((((....)))))))...)))))....((.(((((....--------.))))).))......)))))))))- ( -34.10, z-score = -0.96, R) >droEre2.scaffold_4690 9285789 101 - 18748788 CCUUUCAGCUCACCCAACCCAUUGGGCGGGUGUCCUUCGCAGGACACA--CUAAGCUGCGUGGGCUUUUGGCUUUCGACUAAGCCAACUGUCGACGACAAAAC- .....((((((.(((((....))))).).(((((((....))))))).--...)))))(((.(((..(((((((......)))))))..))).))).......- ( -40.00, z-score = -3.85, R) >droYak2.chrX 17611396 93 - 21770863 --------ACUUUCAGACCCAUUGGGUGGGUGUCCUUCGCAGGACACA--CUAAGCUGCGUGGGCUUUGGGCUUUCGCCUAAGCCAACUGUCGACGACAAAAC- --------........((((((...))))))(((..((((((..(((.--(......).)))(((((.((((....)))))))))..))).))).))).....- ( -32.10, z-score = -1.47, R) >droSec1.super_8 1304402 101 - 3762037 CCUUUCAGCUUCACCACCCCCCUGGGUGGUUGUCCUUCGCAGGACACA--CUAAGCUGCGUGGGCUUUUGGCUUCCAACUAAGCCAACUGUCGACGACAAAAC- .....((((((.(((((((....)))))))((((((....))))))..--..))))))(((.(((..(((((((......)))))))..))).))).......- ( -41.90, z-score = -4.69, R) >droSim1.chrX 14691922 101 - 17042790 CCUUUCAGCUCCACCACCCUCCUGGGUGGUUGUCCUUCGCAGGACACA--CUAAGCUGCGUGGGCUUUUGGCUUUCAACUAAGCCAACUGUCGACGACAAAAC- .....(((((..(((((((....)))))))((((((....))))))..--...)))))(((.(((..(((((((......)))))))..))).))).......- ( -43.10, z-score = -5.26, R) >droMoj3.scaffold_6473 7706860 89 + 16943266 -------GUCUUACAACUCCCCUUGCUCUCUAUGUUUGUGUGUGUGUG--UUUGGGUGCGUUAUGCAGUGGUU------UGUGUUGGUAGUCAACAACGACCUU -------.......(((.((((..((.(.((((......))).).).)--)..))).).))).......((((------..(((((.....)))))..)))).. ( -16.90, z-score = 1.02, R) >consensus ____UCAGCUUCACCAACCCCUUGGGUGGGUGUCCUUCGCAGGACACA__CUAAGCUGCGUGGGCUUUUGGCUUUC_ACUAAGCCAACUGUCGACGACAAAAC_ ..............................((((((....))))))............(((.(((..(.(((..........))).)..))).)))........ (-13.55 = -13.17 + -0.37)
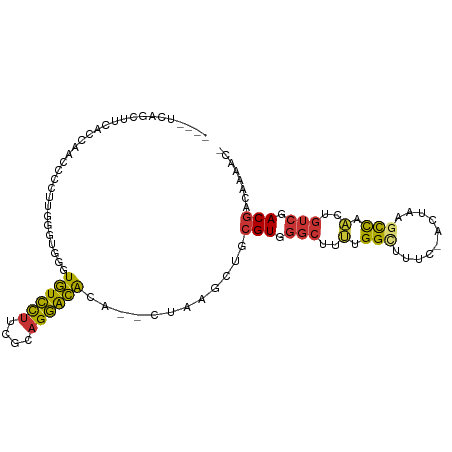
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:16 2011