
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,996,802 – 18,996,869 |
| Length | 67 |
| Max. P | 0.982084 |

| Location | 18,996,802 – 18,996,869 |
|---|---|
| Length | 67 |
| Sequences | 9 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 86.21 |
| Shannon entropy | 0.25151 |
| G+C content | 0.29377 |
| Mean single sequence MFE | -11.34 |
| Consensus MFE | -4.64 |
| Energy contribution | -5.22 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.01 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.982084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
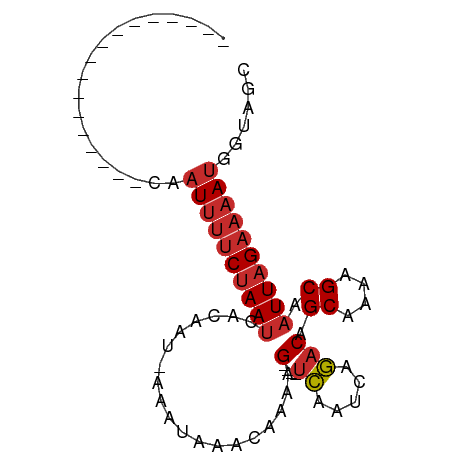
>dm3.chrX 18996802 67 + 22422827 -----------------CAAUUUUCUAAUCACAAU-AAAUAAACAAAAG----UCAAUCAGACAGCAAAAGCAAUUAGAAAAUGGUAGC -----------------..((((((((((......-............(----((.....))).((....)).))))))))))...... ( -11.00, z-score = -4.30, R) >droPer1.super_14 131234 64 + 2168203 --------------------UUUUCUAAUCACAAU-AAAUAAACAAAAG----UCAAUCAGACAGCAAAAGCAAUUAGAAAAUGGUAUC --------------------(((((((((......-............(----((.....))).((....)).)))))))))....... ( -9.50, z-score = -4.26, R) >dp4.chrXL_group1e 10697646 67 + 12523060 -----------------CAAUUUUCUAAUCACAAU-AAAUAAACAAAAG----UCAAUCAGACAGCAAAAGCAAUUAGAAAAUGGUAUC -----------------..((((((((((......-............(----((.....))).((....)).))))))))))...... ( -11.00, z-score = -5.11, R) >droAna3.scaffold_13288 1728 67 - 3652 -----------------CAAUUUUCUAAUCACAAU-AAAUAAACAAAAG----UCAAUCAGACAGCAGAAGCAAUUAGAAAAUGGUAGC -----------------..((((((((((......-............(----((.....))).((....)).))))))))))...... ( -11.00, z-score = -3.76, R) >droYak2.chrX 17595258 67 + 21770863 -----------------CAAUUUUCUAAUCACAAU-AAAUAAACAAAAG----UCAAUCAGACAGCAAAAGCAAUUAGAAAAUGGUAGC -----------------..((((((((((......-............(----((.....))).((....)).))))))))))...... ( -11.00, z-score = -4.30, R) >droEre2.scaffold_4690 9270895 67 + 18748788 -----------------CAAUUUUCUAAUCACAAU-AAAUAAACAAAAG----UCAAUCAGACAGCAAAAGCAAUUAGAAAAUGGUAGC -----------------..((((((((((......-............(----((.....))).((....)).))))))))))...... ( -11.00, z-score = -4.30, R) >droSec1.super_8 1287931 67 + 3762037 -----------------CAAUUUUCUAAUCACAAU-AAAUAAACAAAAG----UCAAUCAGACAGCAAAAGCAAUUAGAAAAUGGUAGC -----------------..((((((((((......-............(----((.....))).((....)).))))))))))...... ( -11.00, z-score = -4.30, R) >droSim1.chrX_random 4928003 67 + 5698898 -----------------CAAUUUUCUAAUCACAAU-AAAUAAACAAAAG----UCAAUCAGACAGCAAAAGCAAUUAGAAAAUGGUAGC -----------------..((((((((((......-............(----((.....))).((....)).))))))))))...... ( -11.00, z-score = -4.30, R) >droMoj3.scaffold_6473 7681727 89 - 16943266 CUAUUUUUGGGUCGCAGAAAUAAACAAAACACAAUCAAGUUGACAAAAGGACGCUGGACAAACAGCAAAAGCAAUUAGAAAAUGGCGGC ((((((((..((.((.......................(((........)))((((......))))....)).))..)))))))).... ( -15.60, z-score = -1.46, R) >consensus _________________CAAUUUUCUAAUCACAAU_AAAUAAACAAAAG____UCAAUCAGACAGCAAAAGCAAUUAGAAAAUGGUAGC ...................((((((((((........................((.....))..((....)).))))))))))...... ( -4.64 = -5.22 + 0.58)

| Location | 18,996,802 – 18,996,869 |
|---|---|
| Length | 67 |
| Sequences | 9 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 86.21 |
| Shannon entropy | 0.25151 |
| G+C content | 0.29377 |
| Mean single sequence MFE | -11.85 |
| Consensus MFE | -7.02 |
| Energy contribution | -7.68 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
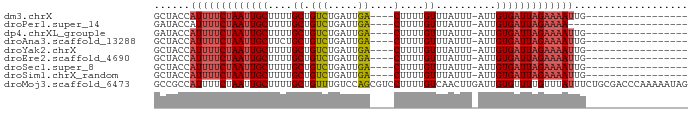
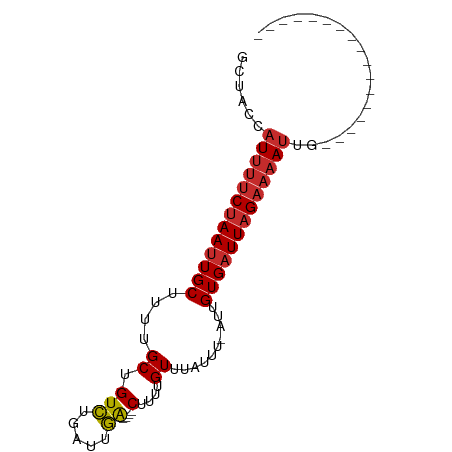
>dm3.chrX 18996802 67 - 22422827 GCUACCAUUUUCUAAUUGCUUUUGCUGUCUGAUUGA----CUUUUGUUUAUUU-AUUGUGAUUAGAAAAUUG----------------- ......((((((((((..(....((.(((.....))----)....))......-...)..))))))))))..----------------- ( -12.42, z-score = -3.55, R) >droPer1.super_14 131234 64 - 2168203 GAUACCAUUUUCUAAUUGCUUUUGCUGUCUGAUUGA----CUUUUGUUUAUUU-AUUGUGAUUAGAAAA-------------------- .......(((((((((..(....((.(((.....))----)....))......-...)..)))))))))-------------------- ( -11.22, z-score = -3.15, R) >dp4.chrXL_group1e 10697646 67 - 12523060 GAUACCAUUUUCUAAUUGCUUUUGCUGUCUGAUUGA----CUUUUGUUUAUUU-AUUGUGAUUAGAAAAUUG----------------- ......((((((((((..(....((.(((.....))----)....))......-...)..))))))))))..----------------- ( -12.42, z-score = -3.64, R) >droAna3.scaffold_13288 1728 67 + 3652 GCUACCAUUUUCUAAUUGCUUCUGCUGUCUGAUUGA----CUUUUGUUUAUUU-AUUGUGAUUAGAAAAUUG----------------- ......((((((((((..(....((.(((.....))----)....))......-...)..))))))))))..----------------- ( -12.42, z-score = -3.43, R) >droYak2.chrX 17595258 67 - 21770863 GCUACCAUUUUCUAAUUGCUUUUGCUGUCUGAUUGA----CUUUUGUUUAUUU-AUUGUGAUUAGAAAAUUG----------------- ......((((((((((..(....((.(((.....))----)....))......-...)..))))))))))..----------------- ( -12.42, z-score = -3.55, R) >droEre2.scaffold_4690 9270895 67 - 18748788 GCUACCAUUUUCUAAUUGCUUUUGCUGUCUGAUUGA----CUUUUGUUUAUUU-AUUGUGAUUAGAAAAUUG----------------- ......((((((((((..(....((.(((.....))----)....))......-...)..))))))))))..----------------- ( -12.42, z-score = -3.55, R) >droSec1.super_8 1287931 67 - 3762037 GCUACCAUUUUCUAAUUGCUUUUGCUGUCUGAUUGA----CUUUUGUUUAUUU-AUUGUGAUUAGAAAAUUG----------------- ......((((((((((..(....((.(((.....))----)....))......-...)..))))))))))..----------------- ( -12.42, z-score = -3.55, R) >droSim1.chrX_random 4928003 67 - 5698898 GCUACCAUUUUCUAAUUGCUUUUGCUGUCUGAUUGA----CUUUUGUUUAUUU-AUUGUGAUUAGAAAAUUG----------------- ......((((((((((..(....((.(((.....))----)....))......-...)..))))))))))..----------------- ( -12.42, z-score = -3.55, R) >droMoj3.scaffold_6473 7681727 89 + 16943266 GCCGCCAUUUUCUAAUUGCUUUUGCUGUUUGUCCAGCGUCCUUUUGUCAACUUGAUUGUGUUUUGUUUAUUUCUGCGACCCAAAAAUAG ...((.(((....))).))....((((......))))((((....((.(((..(((...)))..))).))....).))).......... ( -8.50, z-score = 1.04, R) >consensus GCUACCAUUUUCUAAUUGCUUUUGCUGUCUGAUUGA____CUUUUGUUUAUUU_AUUGUGAUUAGAAAAUUG_________________ ......(((((((((((((......................................)))))))))))))................... ( -7.02 = -7.68 + 0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:11 2011