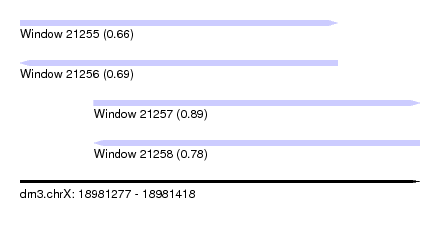
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,981,277 – 18,981,418 |
| Length | 141 |
| Max. P | 0.888947 |

| Location | 18,981,277 – 18,981,389 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 87.96 |
| Shannon entropy | 0.19212 |
| G+C content | 0.42337 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -23.01 |
| Energy contribution | -23.07 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18981277 112 + 22422827 ---CAAAGGAUACGUAAUUUCAGACAGUUCCUGCACACUUCUCUGAGUGGGUAUAUUCAAAGACUCGCAUGCAAGUGCGAAAAUCGCAUCUGCAAAGUAAAUCAAAUCAGAUUAC ---...((((...((........))...)))).........(((((((((((..........)))))).((((..((((.....))))..))))............))))).... ( -26.80, z-score = -2.01, R) >droEre2.scaffold_4690 9255497 115 + 18748788 CCGCCAAGAGUGAGUAAUUUGACACACUUUUUGCACACUUCCCUGAGUGGGCAUAUUCACAUACUCACAUGCCAGUGCGAAAAUCGCAUCUGCAAAGUAAAUCAAAUCAGCUCGU ..((.(((((((.((......)).))))))).))........(((((((((.((......)).))))).(((..(((((.....)))))..)))............))))..... ( -28.80, z-score = -1.66, R) >droSec1.super_8 1272719 112 + 3762037 ---CAAAGGAUACGCAAUUUGACACAGUUCCUGCACACUUCUCUGAGUGGGUGUAUUCAAAGACUCGCAUGCAAGUGCGAAAAUCGCAUCUGCAAAGUAAAUCAAAUCAGAUUAC ---...((((.((.............)))))).........(((((((((((.(......).)))))).((((..((((.....))))..))))............))))).... ( -27.72, z-score = -1.48, R) >droSim1.chrU 1693873 112 - 15797150 ---CAAAGGAUACGUAAUUUGGCACAGUUCCUGCACACUUCUCUGAGUGGGUGUAUUCAAAGACUCGCAUGCAAGUGCGAAAAUCGCAUCUGCAAAGUAAAUCGAAUCAGAUUAC ---..........(((((((((((.......)))..((((......((((((.(......).)))))).((((..((((.....))))..)))))))).........)))))))) ( -28.60, z-score = -1.39, R) >consensus ___CAAAGGAUACGUAAUUUGACACAGUUCCUGCACACUUCUCUGAGUGGGUAUAUUCAAAGACUCGCAUGCAAGUGCGAAAAUCGCAUCUGCAAAGUAAAUCAAAUCAGAUUAC ......((((...((........))...)))).........(((((((((((.(......).)))))).((((.(((((.....))))).))))............))))).... (-23.01 = -23.07 + 0.06)


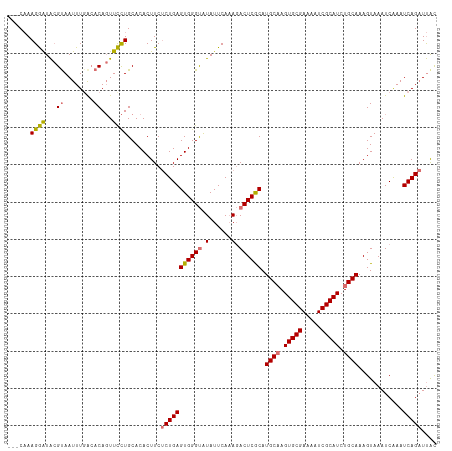
| Location | 18,981,277 – 18,981,389 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 87.96 |
| Shannon entropy | 0.19212 |
| G+C content | 0.42337 |
| Mean single sequence MFE | -32.05 |
| Consensus MFE | -23.21 |
| Energy contribution | -24.71 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.693272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18981277 112 - 22422827 GUAAUCUGAUUUGAUUUACUUUGCAGAUGCGAUUUUCGCACUUGCAUGCGAGUCUUUGAAUAUACCCACUCAGAGAAGUGUGCAGGAACUGUCUGAAAUUACGUAUCCUUUG--- ((((((......)).))))....((((((((.....))).(((((((((...(((((((..........))))))).)))))))))....))))).................--- ( -30.20, z-score = -2.39, R) >droEre2.scaffold_4690 9255497 115 - 18748788 ACGAGCUGAUUUGAUUUACUUUGCAGAUGCGAUUUUCGCACUGGCAUGUGAGUAUGUGAAUAUGCCCACUCAGGGAAGUGUGCAAAAAGUGUGUCAAAUUACUCACUCUUGGCGG ..(((.(((((((((.((((((.(((.((((.....)))))))(((..(..((((....)))).(((.....)))..)..)))..)))))).))))))))))))........... ( -34.90, z-score = -1.53, R) >droSec1.super_8 1272719 112 - 3762037 GUAAUCUGAUUUGAUUUACUUUGCAGAUGCGAUUUUCGCACUUGCAUGCGAGUCUUUGAAUACACCCACUCAGAGAAGUGUGCAGGAACUGUGUCAAAUUGCGUAUCCUUUG--- (((((.((((............((((.((((.....))))(((((((((...(((((((..........))))))).)))))))))..)))))))).)))))..........--- ( -32.90, z-score = -2.44, R) >droSim1.chrU 1693873 112 + 15797150 GUAAUCUGAUUCGAUUUACUUUGCAGAUGCGAUUUUCGCACUUGCAUGCGAGUCUUUGAAUACACCCACUCAGAGAAGUGUGCAGGAACUGUGCCAAAUUACGUAUCCUUUG--- ((((..(((......)))..)))).((((((..(((.((((((((((((...(((((((..........))))))).)))))))).....)))).)))...)))))).....--- ( -30.20, z-score = -2.07, R) >consensus GUAAUCUGAUUUGAUUUACUUUGCAGAUGCGAUUUUCGCACUUGCAUGCGAGUCUUUGAAUACACCCACUCAGAGAAGUGUGCAGGAACUGUGUCAAAUUACGUAUCCUUUG___ ......(((((((((.......((((.((((.....))))((((((..(...(((((((..........))))))).)..))))))..))))))))))))).............. (-23.21 = -24.71 + 1.50)

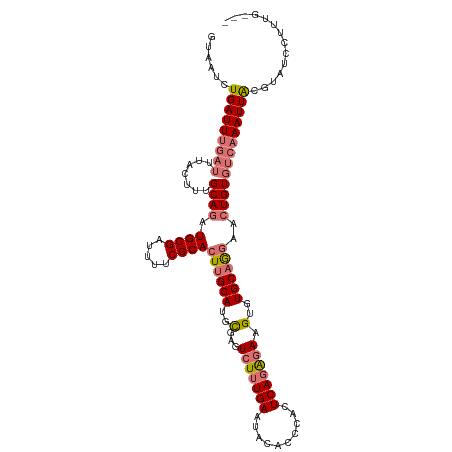
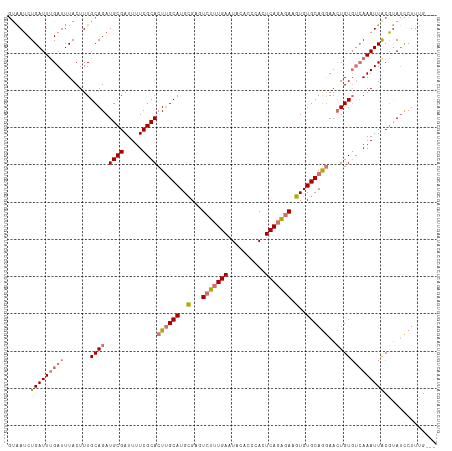
| Location | 18,981,303 – 18,981,418 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 91.16 |
| Shannon entropy | 0.14273 |
| G+C content | 0.48146 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -31.15 |
| Energy contribution | -31.15 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.888947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18981303 115 + 22422827 CCUGCACACUUCUCUGAGUGGGUAUAUUCAAAGACUCGCAUGCAAGUGCGAAAAUCGCAUCUGCAAAGUAAAUCAAAUCAGAUUACGAGCGUCGGGGUGCGGAAAAGUGCGGAUU .((((((...(((.((((((....)))))).))).(((((((((..((((.....))))..)))...((((.((......))))))..........))))))....))))))... ( -36.40, z-score = -2.35, R) >droEre2.scaffold_4690 9255526 114 + 18748788 UUUGCACACUUCCCUGAGUGGGCAUAUUCACAUACUCACAUGCCAGUGCGAAAAUCGCAUCUGCAAAGUAAAUCAAAUCAGCUCGUGA-CGUCGGGGUGCGAAAAAAGGCGGAUU (((((((......(((((((((.((......)).))))).(((..(((((.....)))))..)))............))))((((...-...)))))))))))............ ( -30.60, z-score = -0.73, R) >droSec1.super_8 1272745 115 + 3762037 CCUGCACACUUCUCUGAGUGGGUGUAUUCAAAGACUCGCAUGCAAGUGCGAAAAUCGCAUCUGCAAAGUAAAUCAAAUCAGAUUACGAGCGUCGGGGUGCGGAAAAGUGCGGAUU .((((((..((((((((((((((.(......).)))))).((((..((((.....))))..))))............)))))...((.((......)).)))))..))))))... ( -37.10, z-score = -2.20, R) >droSim1.chrU 1693899 115 - 15797150 CCUGCACACUUCUCUGAGUGGGUGUAUUCAAAGACUCGCAUGCAAGUGCGAAAAUCGCAUCUGCAAAGUAAAUCGAAUCAGAUUACGAGCGUCGGGGUGCGGAAGGGUGCGGAUU .((((((.(((((....((((((.(......).)))))).((((..((((.....))))..))))..(((..((((.((.......))...))))..)))))))).))))))... ( -41.60, z-score = -2.79, R) >consensus CCUGCACACUUCUCUGAGUGGGUAUAUUCAAAGACUCGCAUGCAAGUGCGAAAAUCGCAUCUGCAAAGUAAAUCAAAUCAGAUUACGAGCGUCGGGGUGCGGAAAAGUGCGGAUU .((((((.....(((((((((((.(......).)))))).((((.(((((.....))))).))))............)))))...((.....))..))))))............. (-31.15 = -31.15 + 0.00)


| Location | 18,981,303 – 18,981,418 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 91.16 |
| Shannon entropy | 0.14273 |
| G+C content | 0.48146 |
| Mean single sequence MFE | -30.95 |
| Consensus MFE | -25.58 |
| Energy contribution | -25.32 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18981303 115 - 22422827 AAUCCGCACUUUUCCGCACCCCGACGCUCGUAAUCUGAUUUGAUUUACUUUGCAGAUGCGAUUUUCGCACUUGCAUGCGAGUCUUUGAAUAUACCCACUCAGAGAAGUGUGCAGG ....((((((((((...........((((((((((......)))......(((((.((((.....)))).))))))))))))...(((..........))))))))))))).... ( -31.60, z-score = -2.55, R) >droEre2.scaffold_4690 9255526 114 - 18748788 AAUCCGCCUUUUUUCGCACCCCGACG-UCACGAGCUGAUUUGAUUUACUUUGCAGAUGCGAUUUUCGCACUGGCAUGUGAGUAUGUGAAUAUGCCCACUCAGGGAAGUGUGCAAA ............((((((...(((.(-(((.....)))))))((((((..(((((.((((.....)))))).))).)))))).))))))..(((.((((......)))).))).. ( -27.30, z-score = 0.27, R) >droSec1.super_8 1272745 115 - 3762037 AAUCCGCACUUUUCCGCACCCCGACGCUCGUAAUCUGAUUUGAUUUACUUUGCAGAUGCGAUUUUCGCACUUGCAUGCGAGUCUUUGAAUACACCCACUCAGAGAAGUGUGCAGG ....((((((((((.(((...........((((((......)).))))..(((((.((((.....)))).))))))))((((...((....))...)))).)))))))))).... ( -32.10, z-score = -2.63, R) >droSim1.chrU 1693899 115 + 15797150 AAUCCGCACCCUUCCGCACCCCGACGCUCGUAAUCUGAUUCGAUUUACUUUGCAGAUGCGAUUUUCGCACUUGCAUGCGAGUCUUUGAAUACACCCACUCAGAGAAGUGUGCAGG .....((((.(((((((........))..(((.((.((((((........(((((.((((.....)))).)))))..))))))...)).))).........).)))).))))... ( -32.80, z-score = -2.82, R) >consensus AAUCCGCACUUUUCCGCACCCCGACGCUCGUAAUCUGAUUUGAUUUACUUUGCAGAUGCGAUUUUCGCACUUGCAUGCGAGUCUUUGAAUACACCCACUCAGAGAAGUGUGCAGG .....((((.((((...........((((((((((......)))......(((((.((((.....)))).)))))))))))).(((((..........))))))))).))))... (-25.58 = -25.32 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:07 2011