| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,974,493 – 18,974,592 |
| Length | 99 |
| Max. P | 0.976801 |

| Location | 18,974,493 – 18,974,586 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 66.75 |
| Shannon entropy | 0.58021 |
| G+C content | 0.38484 |
| Mean single sequence MFE | -20.91 |
| Consensus MFE | -9.64 |
| Energy contribution | -10.08 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.795718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18974493 93 + 22422827 --------CAUAAUUAAGCGUG-AUCAUUUCUAU-UAAAGUAAAGUACCUGGUUAUUAAAAUGCCCAGGUAC---------ACCGCCCAUUUGAUAUCCACCACUAGAUGAU----- --------.........(((.(-(.....))...-.........((((((((.(((....))).))))))))---------..))).((((((...........))))))..----- ( -17.70, z-score = -1.77, R) >droSim1.chrU 6263915 89 - 15797150 --------CAUAAUCAAGCGUG-AUCAUUUCUAU-UAAAGUAAAGUACCUGGUUAUUAAAAUGCCCAGGUAC---------ACCGACCAUUUGAUACCCACCACUAGA--------- --------....(((((..(((-.((((((....-..))))...((((((((.(((....))).))))))))---------...)).)))))))).............--------- ( -17.70, z-score = -2.20, R) >droSec1.super_8 1265582 93 + 3762037 --------CAUAAUCAAGCGUG-AUCAUUUCUAU-UAAAGUAAAGUACCUGGUUAUUAAAAUGCCCAGGUAC---------ACCGACCAUUUGAUACCCACCACUAGAUGGU----- --------....((((....))-)).........-.........((((((((.(((....))).))))))))---------....((((((((...........))))))))----- ( -19.70, z-score = -1.72, R) >droYak2.chrX 17575267 115 + 21770863 AGCAAGUUCAUAAUUCAGCGUG-AUCAUUUCUAU-UAAAGUAAAGUACCUGGUUAUUAAGAUGCCCAGGUACGAGUACUACACCACCCAUUCAAUCCCCACCACUGGUGGUGCUGGU ..............(((((...-...........-...((((..((((((((.(((....))).))))))))...))))..((((((..................))))))))))). ( -31.47, z-score = -2.20, R) >droEre2.scaffold_4690 9249001 100 + 18748788 AGGGAAGGCAUAAUGAAGCGUG-ACCACUUCUAU-CAAAGUAAAGUACCUGGUUAUUAAAGUGCCCAGGUAC---------ACCACCCAUUUAAUGGCCACCACUAGCGGU------ .((...(((....((((..(((-...((((....-..))))...((((((((.(((....))).))))))))---------..)))...))))...))).)).........------ ( -25.60, z-score = -0.48, R) >droVir3.scaffold_12472 202306 93 - 763072 ------UAUAUAAAUGUAUGGAUAUUAAAUGAAUAUGAAUUUAAACAUCGCGUCAUUAGAAAGUCAACAUAC-----------CCUAUUUUUAGGGCACAUUAUGAAGGU------- ------......((((.((((((.((((((........))))))..))).)))))))..........(((((-----------((((....))))).....)))).....------- ( -13.30, z-score = -0.04, R) >consensus ________CAUAAUUAAGCGUG_AUCAUUUCUAU_UAAAGUAAAGUACCUGGUUAUUAAAAUGCCCAGGUAC_________ACCGCCCAUUUAAUACCCACCACUAGAGG_______ ............................................((((((((.(((....))).))))))))............................................. ( -9.64 = -10.08 + 0.45)


| Location | 18,974,493 – 18,974,586 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 66.75 |
| Shannon entropy | 0.58021 |
| G+C content | 0.38484 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -13.53 |
| Energy contribution | -12.82 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18974493 93 - 22422827 -----AUCAUCUAGUGGUGGAUAUCAAAUGGGCGGU---------GUACCUGGGCAUUUUAAUAACCAGGUACUUUACUUUA-AUAGAAAUGAU-CACGCUUAAUUAUG-------- -----.(((((....)))))........((((((..---------((((((((............)))))))).........-.((....))..-..))))))......-------- ( -23.20, z-score = -1.95, R) >droSim1.chrU 6263915 89 + 15797150 ---------UCUAGUGGUGGGUAUCAAAUGGUCGGU---------GUACCUGGGCAUUUUAAUAACCAGGUACUUUACUUUA-AUAGAAAUGAU-CACGCUUGAUUAUG-------- ---------((((..((((((.(((........)))---------((((((((............))))))))))))))...-.)))).(((((-((....))))))).-------- ( -22.10, z-score = -1.44, R) >droSec1.super_8 1265582 93 - 3762037 -----ACCAUCUAGUGGUGGGUAUCAAAUGGUCGGU---------GUACCUGGGCAUUUUAAUAACCAGGUACUUUACUUUA-AUAGAAAUGAU-CACGCUUGAUUAUG-------- -----.(((((....)))))..(((((.((((((..---------((((((((............))))))))....((...-..))...))))-))...)))))....-------- ( -25.80, z-score = -2.06, R) >droYak2.chrX 17575267 115 - 21770863 ACCAGCACCACCAGUGGUGGGGAUUGAAUGGGUGGUGUAGUACUCGUACCUGGGCAUCUUAAUAACCAGGUACUUUACUUUA-AUAGAAAUGAU-CACGCUGAAUUAUGAACUUGCU ..((((.(((((...))))).((((....(((((......)))))((((((((............)))))))).........-........)))-)..))))............... ( -32.80, z-score = -1.06, R) >droEre2.scaffold_4690 9249001 100 - 18748788 ------ACCGCUAGUGGUGGCCAUUAAAUGGGUGGU---------GUACCUGGGCACUUUAAUAACCAGGUACUUUACUUUG-AUAGAAGUGGU-CACGCUUCAUUAUGCCUUCCCU ------.((((.....)))).........(((.(((---------((((((((............)))))))).......((-((.((((((..-..)))))))))).)))..))). ( -34.00, z-score = -2.09, R) >droVir3.scaffold_12472 202306 93 + 763072 -------ACCUUCAUAAUGUGCCCUAAAAAUAGG-----------GUAUGUUGACUUUCUAAUGACGCGAUGUUUAAAUUCAUAUUCAUUUAAUAUCCAUACAUUUAUAUA------ -------.......((((((((((((....))))-----------)))))))).......((((....((((((.((((........))))))))))....))))......------ ( -16.50, z-score = -1.77, R) >consensus _______CCUCUAGUGGUGGGUAUCAAAUGGGCGGU_________GUACCUGGGCAUUUUAAUAACCAGGUACUUUACUUUA_AUAGAAAUGAU_CACGCUUAAUUAUG________ ...............((((...((((...................((((((((............))))))))....((......))...))))...))))................ (-13.53 = -12.82 + -0.72)


| Location | 18,974,493 – 18,974,592 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 69.87 |
| Shannon entropy | 0.52625 |
| G+C content | 0.40278 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -12.62 |
| Energy contribution | -14.27 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18974493 99 + 22422827 CAUAAUUAAGCGUGAUCAUU---UCUAU-UAAAGUAAAGUACCUGGUUAUUAAAAUGCCCAGGUAC---------ACCGCCCAUUUGAUAUCCACCACUAGA-----UGAUGGUGCG-- .........(((..((((((---(.(((-((((.....((((((((.(((....))).))))))))---------........))))))).........)))-----))))..))).-- ( -22.52, z-score = -1.87, R) >droSim1.chrU 6263915 89 - 15797150 CAUAAUCAAGCGUGAUCAUU---UCUAU-UAAAGUAAAGUACCUGGUUAUUAAAAUGCCCAGGUAC---------ACCGACCAUUUGAUACCCACCACUAGA----------------- ....(((((..(((.(((((---(....-..))))...((((((((.(((....))).))))))))---------...)).)))))))).............----------------- ( -17.70, z-score = -2.20, R) >droSec1.super_8 1265582 99 + 3762037 CAUAAUCAAGCGUGAUCAUU---UCUAU-UAAAGUAAAGUACCUGGUUAUUAAAAUGCCCAGGUAC---------ACCGACCAUUUGAUACCCACCACUAGA-----UGGUGGUGCG-- ....(((((..(((.(((((---(....-..))))...((((((((.(((....))).))))))))---------...)).))))))))...(((((((...-----.)))))))..-- ( -27.30, z-score = -2.84, R) >droYak2.chrX 17575275 113 + 21770863 CAUAAUUCAGCGUGAUCAUU---UCUAU-UAAAGUAAAGUACCUGGUUAUUAAGAUGCCCAGGUACGAGUACUACACCACCCAUUCAAUCCCCACCACUGGUGGUGCUGGUGGUGCG-- .........(((..((((..---.....-...((((..((((((((.(((....))).))))))))...)))).(((((((..................))))))).))))..))).-- ( -34.07, z-score = -2.19, R) >droEre2.scaffold_4690 9249009 98 + 18748788 CAUAAUGAAGCGUGACCACU---UCUAU-CAAAGUAAAGUACCUGGUUAUUAAAGUGCCCAGGUAC---------ACCACCCAUUUAAUGGCCACCACUAG------CGGUGGUGCG-- .........(((((...(((---(....-..))))...((((((((.(((....))).))))))))---------..)))..........((((((.....------.)))))))).-- ( -29.90, z-score = -2.39, R) >droVir3.scaffold_12472 202306 101 - 763072 UAUAUAAAUGUAUGGAUAUUAAAUGAAUAUGAAUUUAAACAUCGCGUCAUUAGAAAGUCAACAUAC-----------CCUAUUUUUAGGGCACAUUAUGAA-------GGUGGUGUUUG ...................................((((((((((.((((..(.....)......(-----------((((....)))))......)))).-------.)))))))))) ( -17.60, z-score = -0.34, R) >consensus CAUAAUCAAGCGUGAUCAUU___UCUAU_UAAAGUAAAGUACCUGGUUAUUAAAAUGCCCAGGUAC_________ACCGCCCAUUUAAUACCCACCACUAGA_____UGGUGGUGCG__ ......................................((((((((.(((....))).))))))))..........................(((((((.........))))))).... (-12.62 = -14.27 + 1.64)


| Location | 18,974,493 – 18,974,592 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 69.87 |
| Shannon entropy | 0.52625 |
| G+C content | 0.40278 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -13.03 |
| Energy contribution | -13.45 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.959124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18974493 99 - 22422827 --CGCACCAUCA-----UCUAGUGGUGGAUAUCAAAUGGGCGGU---------GUACCUGGGCAUUUUAAUAACCAGGUACUUUACUUUA-AUAGA---AAUGAUCACGCUUAAUUAUG --..((((((..-----....)))))).........((((((..---------((((((((............)))))))).........-.((..---..))....))))))...... ( -27.10, z-score = -2.42, R) >droSim1.chrU 6263915 89 + 15797150 -----------------UCUAGUGGUGGGUAUCAAAUGGUCGGU---------GUACCUGGGCAUUUUAAUAACCAGGUACUUUACUUUA-AUAGA---AAUGAUCACGCUUGAUUAUG -----------------((((..((((((.(((........)))---------((((((((............))))))))))))))...-.))))---.(((((((....))))))). ( -22.10, z-score = -1.44, R) >droSec1.super_8 1265582 99 - 3762037 --CGCACCACCA-----UCUAGUGGUGGGUAUCAAAUGGUCGGU---------GUACCUGGGCAUUUUAAUAACCAGGUACUUUACUUUA-AUAGA---AAUGAUCACGCUUGAUUAUG --.((.((((((-----(...)))))))))(((((.((((((..---------((((((((............))))))))....((...-..)).---..))))))...))))).... ( -30.00, z-score = -2.61, R) >droYak2.chrX 17575275 113 - 21770863 --CGCACCACCAGCACCACCAGUGGUGGGGAUUGAAUGGGUGGUGUAGUACUCGUACCUGGGCAUCUUAAUAACCAGGUACUUUACUUUA-AUAGA---AAUGAUCACGCUGAAUUAUG --.((((((((..(((((((...)))))....))....))))))))((((...((((((((............))))))))..))))...-.....---.................... ( -35.20, z-score = -1.65, R) >droEre2.scaffold_4690 9249009 98 - 18748788 --CGCACCACCG------CUAGUGGUGGCCAUUAAAUGGGUGGU---------GUACCUGGGCACUUUAAUAACCAGGUACUUUACUUUG-AUAGA---AGUGGUCACGCUUCAUUAUG --..((((((..------...))))))((((((.....))))))---------((((((((............)))))))).......((-((.((---((((....)))))))))).. ( -35.70, z-score = -2.96, R) >droVir3.scaffold_12472 202306 101 + 763072 CAAACACCACC-------UUCAUAAUGUGCCCUAAAAAUAGG-----------GUAUGUUGACUUUCUAAUGACGCGAUGUUUAAAUUCAUAUUCAUUUAAUAUCCAUACAUUUAUAUA ...........-------....((((((((((((....))))-----------)))))))).......((((....((((((.((((........))))))))))....))))...... ( -16.50, z-score = -1.55, R) >consensus __CGCACCACCA_____UCUAGUGGUGGGUAUCAAAUGGGCGGU_________GUACCUGGGCAUUUUAAUAACCAGGUACUUUACUUUA_AUAGA___AAUGAUCACGCUUAAUUAUG ....((((((...........))))))..........................((((((((............))))))))...................(((((........))))). (-13.03 = -13.45 + 0.42)

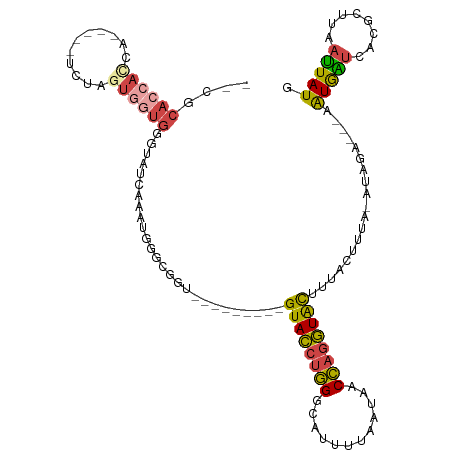

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:59:01 2011