| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,972,940 – 18,973,031 |
| Length | 91 |
| Max. P | 0.945896 |

| Location | 18,972,940 – 18,973,031 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 70.00 |
| Shannon entropy | 0.41260 |
| G+C content | 0.36691 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -13.77 |
| Energy contribution | -14.33 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
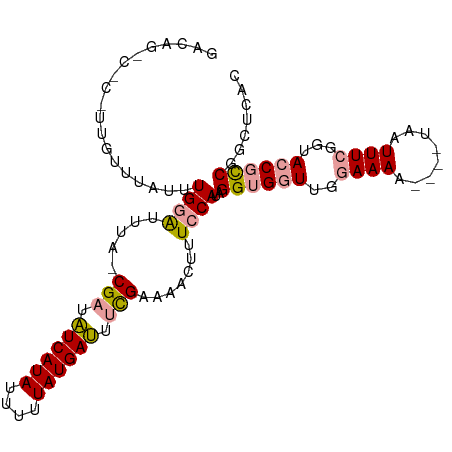
>dm3.chrX 18972940 91 + 22422827 -------------CAUAUUUGCGAUUA-CGAUAUCAUAUUUUUAUGAUUUUGGAAACUUUCCAUAGGUGAUUGGAAAAUAUGUAAUUUCAGUACCGCCGGCUCAC -------------(((((((.((((((-(...((((((....))))))..((((.....))))...)))))))..)))))))....................... ( -17.90, z-score = -1.06, R) >droSim1.chrX 14668153 101 + 17042790 GACAGCCCCAUUGUUUAUAUGGAUUUAUCGAUAUCAUAUUUUUAUGAUUUCGAAAACUUUCCAUAGGUGGUUCGAAAA----UAAUUUCGGUACCGCCGGCUCAC ...((((..........((((((.((.((((.((((((....)))))).)))).))...))))))((((((((((((.----...)))))).))))))))))... ( -35.90, z-score = -5.78, R) >droYak2.chrX 17573646 99 + 21770863 GACAGGCACUUUGUUGAUUUGGAUAUA-CGUUGUCAUAAUUGUAUGACUUCGAAAACUUUCCAUAGGUGGUGGAAAAA----UAGUUUAGAUACC-UCGGCGUAC ............((((((((((((...-((..((((((....))))))..)).....(((((((.....)))))))..----..)))))))....-))))).... ( -19.50, z-score = 0.13, R) >consensus GACAG_C_C_UUGUUUAUUUGGAUUUA_CGAUAUCAUAUUUUUAUGAUUUCGAAAACUUUCCAUAGGUGGUUGGAAAA____UAAUUUCGGUACCGCCGGCUCAC ...................((((.....(((.((((((....)))))).))).......))))..((((((..((((........))))...))))))....... (-13.77 = -14.33 + 0.56)

| Location | 18,972,940 – 18,973,031 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 70.00 |
| Shannon entropy | 0.41260 |
| G+C content | 0.36691 |
| Mean single sequence MFE | -22.40 |
| Consensus MFE | -8.67 |
| Energy contribution | -8.23 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.769307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18972940 91 - 22422827 GUGAGCCGGCGGUACUGAAAUUACAUAUUUUCCAAUCACCUAUGGAAAGUUUCCAAAAUCAUAAAAAUAUGAUAUCG-UAAUCGCAAAUAUG------------- ((((....((((((............((((((((........))))))))........(((((....))))))))))-)..)))).......------------- ( -18.70, z-score = -2.24, R) >droSim1.chrX 14668153 101 - 17042790 GUGAGCCGGCGGUACCGAAAUUA----UUUUCGAACCACCUAUGGAAAGUUUUCGAAAUCAUAAAAAUAUGAUAUCGAUAAAUCCAUAUAAACAAUGGGGCUGUC ...((((((.(((..(((((...----.))))).))).))((((((...((.((((.((((((....)))))).)))).)).))))))..........))))... ( -31.30, z-score = -4.07, R) >droYak2.chrX 17573646 99 - 21770863 GUACGCCGA-GGUAUCUAAACUA----UUUUUCCACCACCUAUGGAAAGUUUUCGAAGUCAUACAAUUAUGACAACG-UAUAUCCAAAUCAACAAAGUGCCUGUC ........(-(((((........----.(((((((.......)))))))....((..((((((....))))))..))-..................))))))... ( -17.20, z-score = -1.25, R) >consensus GUGAGCCGGCGGUACCGAAAUUA____UUUUCCAACCACCUAUGGAAAGUUUUCGAAAUCAUAAAAAUAUGAUAUCG_UAAAUCCAAAUAAACAA_G_G_CUGUC .......((.(((...((((........))))..))).))..((((.....))))..((((((....))))))................................ ( -8.67 = -8.23 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:58:57 2011