
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,965,411 – 18,965,572 |
| Length | 161 |
| Max. P | 0.991485 |

| Location | 18,965,411 – 18,965,572 |
|---|---|
| Length | 161 |
| Sequences | 4 |
| Columns | 166 |
| Reading direction | forward |
| Mean pairwise identity | 62.46 |
| Shannon entropy | 0.53811 |
| G+C content | 0.41341 |
| Mean single sequence MFE | -39.09 |
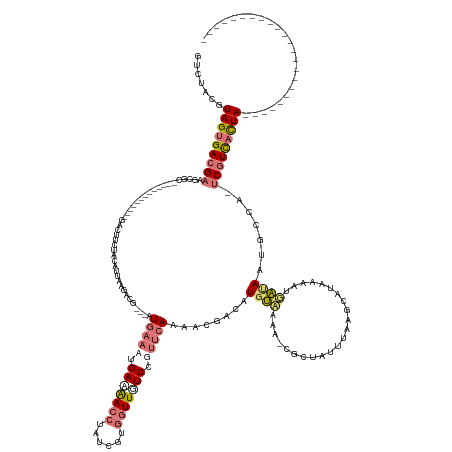
| Consensus MFE | -13.89 |
| Energy contribution | -15.95 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.991485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18965411 161 + 22422827 GUCUACGUAGUGACGAAGCGCACCUGAAGAAGGCGACUUCUGCAUUAAGACGUGCAUUAAAUGAAAACCUAUUGUGGUUGUCCGUUCAUAAUGGCGU-UAUAA-CGCUAUUUAAGCAUAAAAUGAUAAU--CA-GCAUUUUUAUGUUGAAGAUGCCAUCGUGACCA (((.(((..((((((..((((..((..(((((....)))))......))..)))).......((.((((......)))).)))).)))).(((((((-(....-.(((.....)))............(--((-((((....))))))).)))))))))))))).. ( -44.20, z-score = -2.01, R) >droSim1.chrX 14660161 125 + 17042790 GUCUACGUAGUGACGAAGCGC-------------GACUUCUACAUUAAGACG---AUGAAAUGAAAACCUAUCGUGGUUUUCCUUUCAAAACGAUAUGUCAAA-CGCUAUUUAAUCAUAAAAUGAUAAUGCCA-UCGUCACUA----------------------- .......(((((((((((((.-------------(((.(((......)))..---.(((((.(((((((......))))))).))))).........)))...-)))).....(((((...))))).......-)))))))))----------------------- ( -33.50, z-score = -3.94, R) >droSec1.super_8 1256451 125 + 3762037 GUCUACGUAGUGACGAAGCGC-------------GACUUCUACAUUCAGACG---AUGAAAUGAAAACCUAUCGUGGUUGUCCUUUCAAAACGAUAUGUCAAA-CGCUAUUUAAGCAUAAAAUGAUAAUGCCU-UCGUCACUA----------------------- .......(((((((((((.((-------------(((.(((......)))..---.(((((.((.((((......)))).)).))))).........)))...-.(((.....))).............))))-)))))))))----------------------- ( -34.50, z-score = -3.48, R) >droEre2.scaffold_4690 9240342 129 + 18748788 GACCGCGUAGUGACGAAGCGC--------------GCCCGAGGAAGGCGACUAACAUGUGAAGACGAAGAAAUGUGGUUGUCCGAUCACAACGACAUGCAUUUUCGGGAGAAAAUGAAAAUGUUGCAAGGGCAAUCGUCACUA----------------------- .......(((((((((.....--------------((((((....((((((((.(((..............)))))))))))...))....((((((.(((((((....)))))))...))))))...))))..)))))))))----------------------- ( -44.14, z-score = -2.92, R) >consensus GUCUACGUAGUGACGAAGCGC_____________GACUUCUACAUUAAGACG___AUGAAAUGAAAACCUAUCGUGGUUGUCCGUUCAAAACGACAUGUAAAA_CGCUAUUUAAGCAUAAAAUGAUAAUGCCA_UCGUCACUA_______________________ .......(((((((((........................................(((((.(((((((......))))))).)))))........((((.......................)))).......)))))))))....................... (-13.89 = -15.95 + 2.06)

| Location | 18,965,411 – 18,965,572 |
|---|---|
| Length | 161 |
| Sequences | 4 |
| Columns | 166 |
| Reading direction | reverse |
| Mean pairwise identity | 62.46 |
| Shannon entropy | 0.53811 |
| G+C content | 0.41341 |
| Mean single sequence MFE | -37.12 |
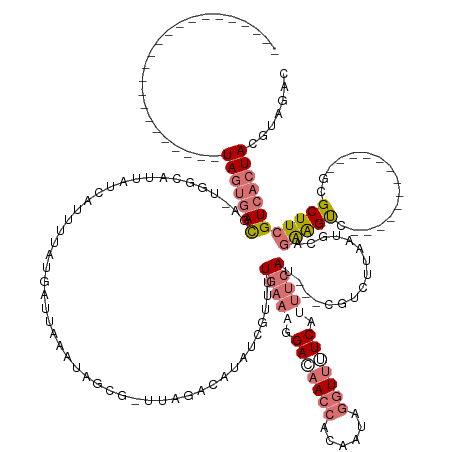
| Consensus MFE | -13.23 |
| Energy contribution | -16.85 |
| Covariance contribution | 3.62 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18965411 161 - 22422827 UGGUCACGAUGGCAUCUUCAACAUAAAAAUGC-UG--AUUAUCAUUUUAUGCUUAAAUAGCG-UUAUA-ACGCCAUUAUGAACGGACAACCACAAUAGGUUUUCAUUUAAUGCACGUCUUAAUGCAGAAGUCGCCUUCUUCAGGUGCGCUUCGUCACUACGUAGAC ...(((..(((((....(((.(((....))).-))--)..........(((((.....))))-)....-..)))))..)))(((((.((((......)))).))......((((........))))((((((((((.....))))).))))).......))).... ( -38.50, z-score = -1.60, R) >droSim1.chrX 14660161 125 - 17042790 -----------------------UAGUGACGA-UGGCAUUAUCAUUUUAUGAUUAAAUAGCG-UUUGACAUAUCGUUUUGAAAGGAAAACCACGAUAGGUUUUCAUUUCAU---CGUCUUAAUGUAGAAGUC-------------GCGCUUCGUCACUACGUAGAC -----------------------(((((((((-.(((...(((((...)))))......(((-(((.((((..((...(((((.(((((((......))))))).))))).---)).....))))..))).)-------------))))))))))))))....... ( -36.70, z-score = -3.54, R) >droSec1.super_8 1256451 125 - 3762037 -----------------------UAGUGACGA-AGGCAUUAUCAUUUUAUGCUUAAAUAGCG-UUUGACAUAUCGUUUUGAAAGGACAACCACGAUAGGUUUUCAUUUCAU---CGUCUGAAUGUAGAAGUC-------------GCGCUUCGUCACUACGUAGAC -----------------------(((((((((-((((...........(((((.....))))-)..(((....((...(((((.((.((((......)))).)).))))).---))((((....)))).)))-------------)).)))))))))))....... ( -37.50, z-score = -3.38, R) >droEre2.scaffold_4690 9240342 129 - 18748788 -----------------------UAGUGACGAUUGCCCUUGCAACAUUUUCAUUUUCUCCCGAAAAUGCAUGUCGUUGUGAUCGGACAACCACAUUUCUUCGUCUUCACAUGUUAGUCGCCUUCCUCGGGC--------------GCGCUUCGUCACUACGCGGUC -----------------------(((((((((..((......(((((...(((((((....))))))).((((.(((((......))))).))))..............))))).(.(((((.....))))--------------)))).)))))))))....... ( -35.80, z-score = -2.08, R) >consensus _______________________UAGUGACGA_UGGCAUUAUCAUUUUAUGAUUAAAUAGCG_UUAGACAUAUCGUUUUGAAAGGACAACCACAAUAGGUUUUCAUUUCAU___CGUCUUAAUGCAGAAGUC_____________GCGCUUCGUCACUACGUAGAC .......................(((((((................................................(((((.(((((((......))))))).)))))................((((((((((.....))))).))))))))))))....... (-13.23 = -16.85 + 3.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:58:56 2011