
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,950,197 – 11,950,318 |
| Length | 121 |
| Max. P | 0.729142 |

| Location | 11,950,197 – 11,950,297 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.11 |
| Shannon entropy | 0.40675 |
| G+C content | 0.49334 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -16.01 |
| Energy contribution | -15.21 |
| Covariance contribution | -0.79 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.729142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
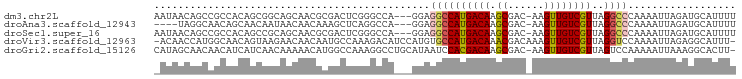
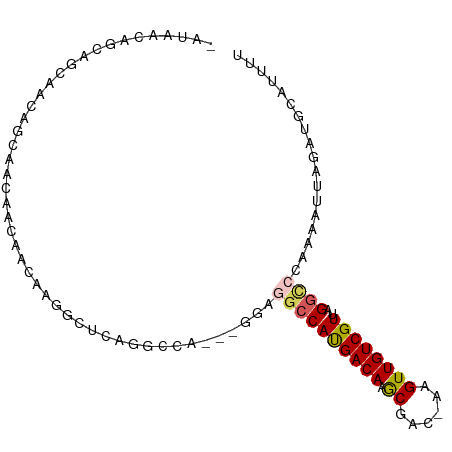
>dm3.chr2L 11950197 100 + 23011544 UCUCGCCUGCGAUCUAUGCCUGGCAUACAAAAUGCAUCUAAUUUUGGGCCUAACGACAACUU-GUCGCUUGUCAUGGCCUCCUGGCCCGAGUCGCGUUGCU---- ....((.(((((.((..(((.((....((((((.......))))))((((....(((((...-.....)))))..)))).)).)))...)))))))..)).---- ( -29.90, z-score = -1.54, R) >droPer1.super_1 1511643 92 - 10282868 --------UCUCUCUCUGUUUGGCAUACAAAAUGCAUCUAAUUUUGGGCCUAACGACAACUU-GUCGCUUGUCAUGGUCUCCCGGUCUGAUCUAUGUUGCU---- --------.........(((.(((...((((((.......)))))).))).)))(((.....-)))((....(((((((.........)).)))))..)).---- ( -20.10, z-score = -1.58, R) >dp4.chr4_group3 4366224 92 - 11692001 --------UCUCUCUCUGUUUGGCAUACAAAAUGCACCUAAUUUUGGGCCUAACGACAACUU-GUCGCUUGUCAUGGUCUCCCGGUCUGAUCUAUGUUGCU---- --------.........(((.(((...((((((.......)))))).))).)))(((.....-)))((....(((((((.........)).)))))..)).---- ( -20.10, z-score = -1.37, R) >droAna3.scaffold_12943 2746420 100 - 5039921 UCUGGCCCGCGAUCUGUGCCAGGCAUACAAAAUGCAUCUAAUUUUGGGCCUAACGACAACUU-GUCGCUUGUCAUGGCCUCCUGGCCUGAGCUUUGUUGUU---- ...((((((((.....)))...((((.....))))..........)))))....((((((..-...((((.....((((....)))).))))...))))))---- ( -30.50, z-score = -1.19, R) >droEre2.scaffold_4929 13181010 104 - 26641161 UCUCGCCAGCGAUCUAUGCCUGGCAUACAAAAUGCAUCUAAUUUUGGGCCUAACGACAACUU-GUCGCUUGUCAUGGCCUCCUGGCCCGAGUCGCGUUGUUGUAG ....(((((((.....)).))))).((((((((((..((.....((((((....(((.....-)))(((......))).....))))))))..))))).))))). ( -32.60, z-score = -1.88, R) >droYak2.chr2L 8371278 100 + 22324452 UCUCGCCAGCGAUCUAUGCCUGGCAUACAAAAUGCAUCUAAUUUUGGGCCUAACGACAACUU-GUCGCUUGUCAUGGCCUCCUGGCCCGAGUCGCGUUGUU---- ....(((((((.....)).)))))......(((((..((.....((((((....(((.....-)))(((......))).....))))))))..)))))...---- ( -28.90, z-score = -1.05, R) >droSec1.super_16 149415 100 + 1878335 UCUCGCCUGCGAUCCAUGCCUGGCAUACAAAAUGCAUCUAAUUUUGGGCCUAACGACAACUU-GUCGCUUGUCAUGGCCUCCUGGCCCGAGUCGCGUUGCU---- ....((.(((((..(..(((.((....((((((.......))))))((((....(((((...-.....)))))..)))).)).)))..)..)))))..)).---- ( -30.20, z-score = -1.43, R) >droVir3.scaffold_12963 6968751 87 - 20206255 ---------------AUGCC-GGCAUGUAAA-UGCCUCUAAUUUUGGACCUAACGACAACUUUGUCGUUUGUCAUGGCACAUGGAUGUCUUUGGCAUUGUUGUU- ---------------..(((-(((((....)-))))..........(((..(((((((....))))))).)))..)))(((..(((((.....)))))..))).- ( -29.10, z-score = -3.28, R) >consensus UCUCGCC_GCGAUCUAUGCCUGGCAUACAAAAUGCAUCUAAUUUUGGGCCUAACGACAACUU_GUCGCUUGUCAUGGCCUCCUGGCCCGAGUCGCGUUGCU____ ................(((...((((.....))))........(((((((....(((......)))(((......))).....)))))))...)))......... (-16.01 = -15.21 + -0.79)



| Location | 11,950,225 – 11,950,318 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 71.56 |
| Shannon entropy | 0.49923 |
| G+C content | 0.46915 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -9.86 |
| Energy contribution | -10.22 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2L 11950225 93 - 23011544 AAUAACAGCCGCCACAGCGGCAGCAACGCGACUCGGGCCA---GGAGGCCAUGACAAGCGAC-AAGUUGUCGUUAGGCCCAAAAUUAGAUGCAUUUU .......(((((....))))).(((.(.......(((((.---((...))......((((((-.....)))))).))))).......).)))..... ( -31.24, z-score = -2.35, R) >droAna3.scaffold_12943 2746448 89 + 5039921 ----UAGGCAACAGCAACAAUAACAACAAAGCUCAGGCCA---GGAGGCCAUGACAAGCGAC-AAGUUGUCGUUAGGCCCAAAAUUAGAUGCAUUUU ----..(((.......................(((((((.---...)))).)))(.((((((-.....)))))).)))).................. ( -20.60, z-score = -1.58, R) >droSec1.super_16 149443 93 - 1878335 AAUAACAGCCGCCACAGCCGCAGCAACGCGACUCGGGCCA---GGAGGCCAUGACAAGCGAC-AAGUUGUCGUUAGGCCCAAAAUUAGAUGCAUUUU .......((.((....)).)).(((.(.......(((((.---((...))......((((((-.....)))))).))))).......).)))..... ( -25.14, z-score = -0.54, R) >droVir3.scaffold_12963 6968763 95 + 20206255 -ACAACCAUGGCAACAGUAAGAACAACAAUGCCAAAGACAUCCAUGUGCCAUGACAAACGACAAAGUUGUCGUUAGGUCCAAAAUUAGAGGCAUUU- -.......(((((...((.......))..)))))...........(((((..(((.(((((((....)))))))..)))(.......).)))))..- ( -19.50, z-score = -1.14, R) >droGri2.scaffold_15126 6618765 95 - 8399593 CAUAGCAACAACAUCAUCAACAAAAACAUGGCCAAAGGCCUGCAUAAUCCACGACAAGCGAC-AAGUUGUCGUUAGUCCAAAAAUUAAAGGCACUU- ....((....................((.((((...))))))..........(((.((((((-.....)))))).)))............))....- ( -18.90, z-score = -2.37, R) >consensus _AUAACAGCAGCAACAGCAACAACAACAAGGCUCAGGCCA___GGAGGCCAUGACAAGCGAC_AAGUUGUCGUUAGGCCCAAAAUUAGAUGCAUUUU ..............................................((((((((((.((......))))))))..)))).................. ( -9.86 = -10.22 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:58 2011