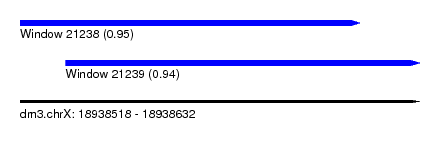
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,938,518 – 18,938,632 |
| Length | 114 |
| Max. P | 0.950179 |

| Location | 18,938,518 – 18,938,615 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.90 |
| Shannon entropy | 0.51562 |
| G+C content | 0.44664 |
| Mean single sequence MFE | -27.69 |
| Consensus MFE | -15.43 |
| Energy contribution | -15.70 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
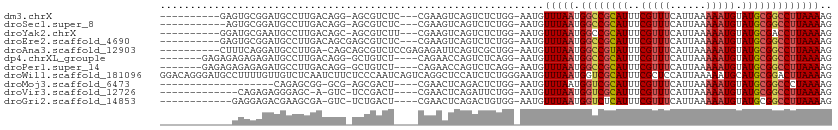
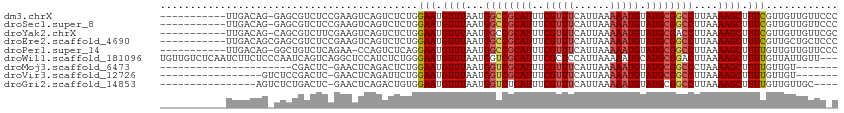
>dm3.chrX 18938518 97 + 22422827 ----------GAGUGCGGAUGCCUUGACAGG-AGCGUCUC---CGAAGUCAGUCUCUGG-AAUGUUUAAUGGCCGCAUUUCGUUUCAUUAAAAAUGUAUGCGGCCUUAAAAG ----------.....((((.((..((((.((-((...)))---)...)))))).)))).-....(((((.((((((((..(((((......))))).))))))))))))).. ( -31.30, z-score = -2.53, R) >droSec1.super_8 1232240 96 + 3762037 -----------AGUGCGGAUGCCUUGACAGG-AGCGUCUC---CGAAGUCAGUCUCUGG-AAUGUUUAAUGGCCGCAUUUCGUUUCAUUAAAAAUGUAUGCGGCCUUAAAAG -----------....((((.((..((((.((-((...)))---)...)))))).)))).-....(((((.((((((((..(((((......))))).))))))))))))).. ( -31.30, z-score = -2.67, R) >droYak2.chrX 17544737 97 + 21770863 ----------GGAUGCGAAUGCCUUGACAGC-AGCGUCUU---CGAAGUCAGUCUCUGG-AAUGUUUAAUGGCCGCAUUUCGUUUCAUUAAAAAUGUAUGCGACCUUAAAAG ----------(((((((...(((..((((.(-((.(.((.---.......)).).))).-..))))....)))))))).((((..(((.....)))...))))))....... ( -20.30, z-score = 0.35, R) >droEre2.scaffold_4690 9221063 98 + 18748788 ----------GAGUGCGGAUGCCUUGACAGCGAGCGUCUC---CGAAGUCAGUCUCUGG-AAUGUUUAAUGGCCGCAUUUCGUUUCAUUAAAAAUGUAUGCGGCCUUAAAAG ----------((.(.((((.((((((....)))).)).))---)).).)).........-....(((((.((((((((..(((((......))))).))))))))))))).. ( -31.50, z-score = -2.40, R) >droAna3.scaffold_12903 14713 100 - 802071 ----------CUUUCAGGAUGCCUUGA-CAGCAGCGUCUCCGAGAGAUUCAGUCGCUGG-AAUGUUUAAUGGCCGUAUUUCGUUUCAUUAAAAAUGUAUGCGGCCUUAAAAG ----------...(((((....)))))-((((.(((((((...)))))...)).)))).-....(((((.((((((((..(((((......))))).))))))))))))).. ( -30.20, z-score = -2.42, R) >dp4.chrXL_group1e 10646163 99 + 12523060 -------GAGAGAGAGAGAUGCCUUGACAGG-GCUGUCU----CAGAACCAGUCUCAGG-AAUGUUUAAUGGCCGCAUUUCGUUUCAUUAAAAAUGUAUGCGGCCUUAAAAG -------....(((((((((((((.....))-)).))))----)........))))...-....(((((.((((((((..(((((......))))).))))))))))))).. ( -32.50, z-score = -2.91, R) >droPer1.super_14 79857 99 + 2168203 -------GAGAGAGAGAGAUGCCUUGACAGG-GCUGUCU----CAGAACCAGUCUCAGG-AAUGUUUAAUGGCCGCAUUUCGUUUCAUUAAAAAUGUAUGCGGCCUUAAAAG -------....(((((((((((((.....))-)).))))----)........))))...-....(((((.((((((((..(((((......))))).))))))))))))).. ( -32.50, z-score = -2.91, R) >droWil1.scaffold_181096 4257054 112 - 12416693 GGACAGGGAUGCCUUUUGUUGUCUCAAUCUUCUCCCAAUCAGUCAGGCUCCAUCUCUGGGAAUGUUUAAUGGUCGCAUUUCGCUCCAUUAAAAAUGCAUGCGGACUUAAAAG .((..((((......(((......))).....))))..))((((..((((((....)))).(((((((((((..((.....)).)))))))....)))))).))))...... ( -23.20, z-score = 0.61, R) >droMoj3.scaffold_6473 7616476 86 - 16943266 -------------------CAGAGCGG-GCG-AGCGACU----CGAACUCAGACUCUGG-AAUGUUUAAUGGUCGCAUUUCGUUUCAUUAAAAAUGUAUGCGGCCCUAAAAG -------------------((((((((-(((-((...))----))..))).).))))).-..........((((((((..(((((......))))).))))))))....... ( -24.70, z-score = -1.66, R) >droVir3.scaffold_12726 880714 91 + 2840439 -------------CAGAGAGGGAGC-A-GUC-UCCGACU----CGAACUCAGAUUCUGG-AAUGUUUAAUGGUCGCAUUUCGUUUCAUUAAAAAUGUAUGCGGCCUUAAAAG -------------(((((((((((.-.-..)-)))..))----)(....)....)))).-....(((((.((((((((..(((((......))))).))))))))))))).. ( -25.10, z-score = -1.83, R) >droGri2.scaffold_14853 5313460 93 + 10151454 ------------GAGGAGACGAAGCGA-GUC-UCUGACU----CGAACUCAGACUGUGG-AAUGUUUAAUGGUCUCAUUUCGUUUCAUUAAAAAUGUAUGCGGCCUUAAAAG ------------...((((((((((((-(((-...))))----)).....(((((((..-........)))))))..))))))))).......................... ( -22.00, z-score = -0.59, R) >consensus ____________GUGCGGAUGCCUUGACAGG_AGCGUCU____CGAACUCAGUCUCUGG_AAUGUUUAAUGGCCGCAUUUCGUUUCAUUAAAAAUGUAUGCGGCCUUAAAAG ................................................................(((((.((((((((..(((((......))))).))))))))))))).. (-15.43 = -15.70 + 0.27)
| Location | 18,938,531 – 18,938,632 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.37 |
| Shannon entropy | 0.41781 |
| G+C content | 0.41470 |
| Mean single sequence MFE | -24.63 |
| Consensus MFE | -16.47 |
| Energy contribution | -16.58 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18938531 101 + 22422827 -----------UUGACAG-GAGCGUCUCCGAAGUCAGUCUCUGGAAUGUUUAAUGGCCGCAUUUCGUUUCAUUAAAAAUGUAUGCGGCCUUAAAAGCUUUCGUUGUUGUUCCC -----------......(-(((((.(..(((((((((...))))....(((((.((((((((..(((((......))))).)))))))))))))...)))))..).)))))). ( -31.60, z-score = -3.55, R) >droSec1.super_8 1232252 101 + 3762037 -----------UUGACAG-GAGCGUCUCCGAAGUCAGUCUCUGGAAUGUUUAAUGGCCGCAUUUCGUUUCAUUAAAAAUGUAUGCGGCCUUAAAAGCUUUCGUUGUUGUUCCC -----------......(-(((((.(..(((((((((...))))....(((((.((((((((..(((((......))))).)))))))))))))...)))))..).)))))). ( -31.60, z-score = -3.55, R) >droYak2.chrX 17544750 101 + 21770863 -----------UUGACAG-CAGCGUCUUCGAAGUCAGUCUCUGGAAUGUUUAAUGGCCGCAUUUCGUUUCAUUAAAAAUGUAUGCGACCUUAAAAGCUUUCGUUGUUGUUCGC -----------..(((((-(((((.....((((((((...))).....(((((.((.(((((..(((((......))))).))))).))))))).)))))))))))))))... ( -25.70, z-score = -1.49, R) >droEre2.scaffold_4690 9221076 102 + 18748788 -----------UUGACAGCGAGCGUCUCCGAAGUCAGUCUCUGGAAUGUUUAAUGGCCGCAUUUCGUUUCAUUAAAAAUGUAUGCGGCCUUAAAAGCUUUCGUUGCUGCUCCC -----------..((((((.((((..((((.((.....)).))))...(((((.((((((((..(((((......))))).)))))))))))))......)))))))).)).. ( -32.10, z-score = -2.98, R) >droPer1.super_14 79873 100 + 2168203 -----------UUGACAG-GGCUGUCUCAGAA-CCAGUCUCAGGAAUGUUUAAUGGCCGCAUUUCGUUUCAUUAAAAAUGUAUGCGGCCUUAAAAGCUUUUGUUGUUGUUCCC -----------..(((.(-(.(((...)))..-)).)))...(((((.(((((.((((((((..(((((......))))).)))))))))))))(((....)))...))))). ( -28.80, z-score = -2.41, R) >droWil1.scaffold_181096 4257070 110 - 12416693 UGUUGUCUCAAUCUUCUCCCAAUCAGUCAGGCUCCAUCUCUGGGAAUGUUUAAUGGUCGCAUUUCGCUCCAUUAAAAAUGCAUGCGGACUUAAAAGCUUUUGUUAUUGUU--- ....((((((......(((((...((...(....)..)).)))))...((((((((..((.....)).))))))))......)).)))).....................--- ( -17.50, z-score = 0.80, R) >droMoj3.scaffold_6473 7616489 83 - 16943266 ----------------------CGACUC-GAACUCAGACUCUGGAAUGUUUAAUGGUCGCAUUUCGUUUCAUUAAAAAUGUAUGCGGCCCUAAAAGCUUUUGUUGU------- ----------------------((((..-.....(((...)))(((.((((...((((((((..(((((......))))).))))))))....)))).))))))).------- ( -18.00, z-score = -1.07, R) >droVir3.scaffold_12726 880727 88 + 2840439 -----------------GUCUCCGACUC-GAACUCAGAUUCUGGAAUGUUUAAUGGUCGCAUUUCGUUUCAUUAAAAAUGUAUGCGGCCUUAAAAGCUUUUGUUGU------- -----------------...(((((.((-.......)).)).)))...(((((.((((((((..(((((......))))).)))))))))))))............------- ( -20.60, z-score = -1.92, R) >droGri2.scaffold_14853 5313474 92 + 10151454 ----------------AGUCUCUGACUC-GAACUCAGACUGUGGAAUGUUUAAUGGUCUCAUUUCGUUUCAUUAAAAAUGUAUGCGGCCUUAAAAGCUUUUGUUGUUGC---- ----------------((((...)))).-.......(((.(..(((..(((((.((((.(((..(((((......))))).))).)))))))))...)))..).)))..---- ( -15.80, z-score = 0.16, R) >consensus ___________UUGACAG_CAGCGUCUCCGAAGUCAGUCUCUGGAAUGUUUAAUGGCCGCAUUUCGUUUCAUUAAAAAUGUAUGCGGCCUUAAAAGCUUUUGUUGUUGUUC_C ...........................................(((.((((...((((((((..(((((......))))).))))))))....)))).)))............ (-16.47 = -16.58 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:58:52 2011