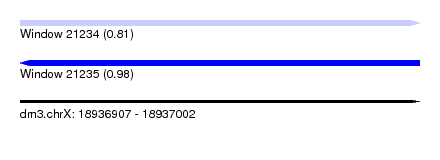
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,936,907 – 18,937,002 |
| Length | 95 |
| Max. P | 0.980129 |

| Location | 18,936,907 – 18,937,002 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 82.79 |
| Shannon entropy | 0.29320 |
| G+C content | 0.32320 |
| Mean single sequence MFE | -11.22 |
| Consensus MFE | -10.00 |
| Energy contribution | -10.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.807170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
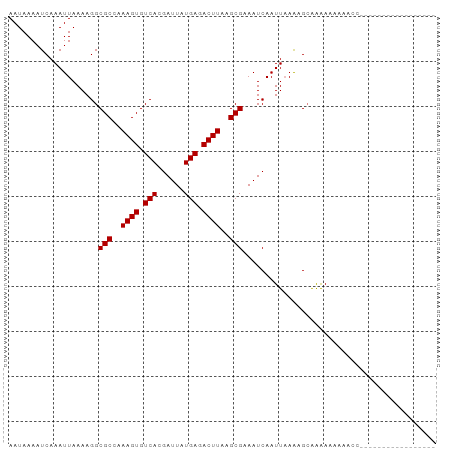
>dm3.chrX 18936907 95 + 22422827 AAUAAAAUCAAAUUAAAAGGCGCCAAAGUGUCACGAUUAUGAGACUUAAGCGAAAUCAAUUAAAAGCAAAAAAGCUGUGGCGAAAAAAAACCAGC ....................(((((((((.(((......))).)))).................(((......))).)))))............. ( -15.80, z-score = -1.34, R) >droSim1.chrX_random 4902601 82 + 5698898 AAUAAAAUCAAAUUAAAAGGCGCCAAAGUGUCACGAUUAUGAGACUUAAGCGAAAUCAAUUAAAAGCAGAAAAAAAACCAGC------------- ....................(((..((((.(((......))).))))..)))..............................------------- ( -10.00, z-score = -1.20, R) >droSec1.super_8 1230720 81 + 3762037 AAUAAAAUCAAAUUAAAAGGCGCCAAAGUGUCACGAUUAUGAGACUUAAGCGAAAUCAAUUUAAAGCAAAAAAAAACCAGC-------------- .........(((((......(((..((((.(((......))).))))..))).....)))))...................-------------- ( -10.30, z-score = -1.23, R) >droEre2.scaffold_4690 9219721 77 + 18748788 AAUAAAAUCAAAUUAAAAGGCGCCAAAGUGUCACGAUUAUGAGACUUAAGCGAAAUCAAUUAAG-GCAAAAAAACCCC----------------- ....................(((..((((.(((......))).))))..)))............-.............----------------- ( -10.00, z-score = -1.17, R) >droAna3.scaffold_12903 13615 76 - 802071 AAUAAAAUCAAAUUAAAAGGCGCCAAAGUGUCACGAUUAUGAGACUUAAGCGAAAUCAAUUAAAAGGGAGAGAAAA------------------- ....................(((..((((.(((......))).))))..)))........................------------------- ( -10.00, z-score = -1.03, R) >consensus AAUAAAAUCAAAUUAAAAGGCGCCAAAGUGUCACGAUUAUGAGACUUAAGCGAAAUCAAUUAAAAGCAAAAAAAAACC_________________ ....................(((..((((.(((......))).))))..)))........................................... (-10.00 = -10.00 + 0.00)

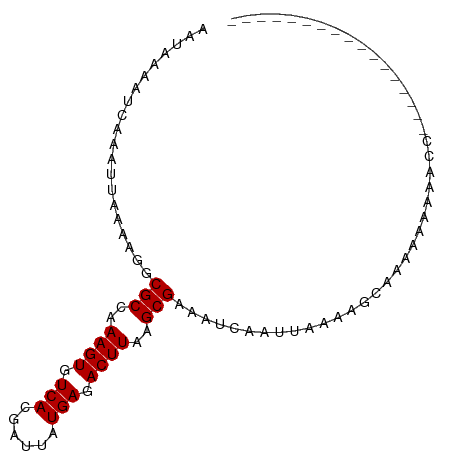
| Location | 18,936,907 – 18,937,002 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 82.79 |
| Shannon entropy | 0.29320 |
| G+C content | 0.32320 |
| Mean single sequence MFE | -15.30 |
| Consensus MFE | -12.62 |
| Energy contribution | -12.62 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
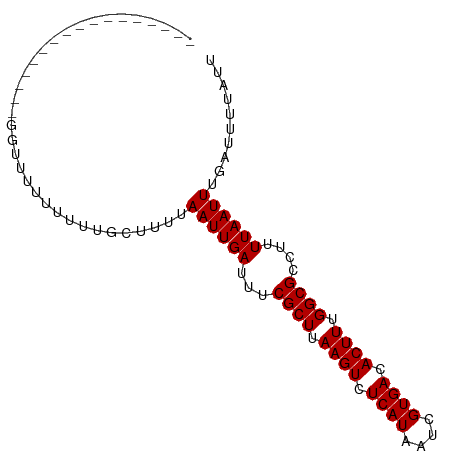
>dm3.chrX 18936907 95 - 22422827 GCUGGUUUUUUUUCGCCACAGCUUUUUUGCUUUUAAUUGAUUUCGCUUAAGUCUCAUAAUCGUGACACUUUGGCGCCUUUUAAUUUGAUUUUAUU ..((((........)))).(((......)))((.((((((...((((.((((.((((....)))).)))).))))....)))))).))....... ( -17.20, z-score = -1.59, R) >droSim1.chrX_random 4902601 82 - 5698898 -------------GCUGGUUUUUUUUCUGCUUUUAAUUGAUUUCGCUUAAGUCUCAUAAUCGUGACACUUUGGCGCCUUUUAAUUUGAUUUUAUU -------------((.((.......)).)).((.((((((...((((.((((.((((....)))).)))).))))....)))))).))....... ( -13.60, z-score = -1.68, R) >droSec1.super_8 1230720 81 - 3762037 --------------GCUGGUUUUUUUUUGCUUUAAAUUGAUUUCGCUUAAGUCUCAUAAUCGUGACACUUUGGCGCCUUUUAAUUUGAUUUUAUU --------------((..(......)..)).(((((((((...((((.((((.((((....)))).)))).))))....)))))))))....... ( -16.40, z-score = -2.70, R) >droEre2.scaffold_4690 9219721 77 - 18748788 -----------------GGGGUUUUUUUGC-CUUAAUUGAUUUCGCUUAAGUCUCAUAAUCGUGACACUUUGGCGCCUUUUAAUUUGAUUUUAUU -----------------(((((......))-)))((((((...((((.((((.((((....)))).)))).))))....)))))).......... ( -16.60, z-score = -2.39, R) >droAna3.scaffold_12903 13615 76 + 802071 -------------------UUUUCUCUCCCUUUUAAUUGAUUUCGCUUAAGUCUCAUAAUCGUGACACUUUGGCGCCUUUUAAUUUGAUUUUAUU -------------------............((.((((((...((((.((((.((((....)))).)))).))))....)))))).))....... ( -12.70, z-score = -2.99, R) >consensus _________________GGUUUUUUUUUGCUUUUAAUUGAUUUCGCUUAAGUCUCAUAAUCGUGACACUUUGGCGCCUUUUAAUUUGAUUUUAUU ..................................((((((...((((.((((.((((....)))).)))).))))....)))))).......... (-12.62 = -12.62 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:58:49 2011