| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,941,263 – 11,941,356 |
| Length | 93 |
| Max. P | 0.570430 |

| Location | 11,941,263 – 11,941,356 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 64.94 |
| Shannon entropy | 0.61520 |
| G+C content | 0.47414 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -9.33 |
| Energy contribution | -8.99 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.570430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
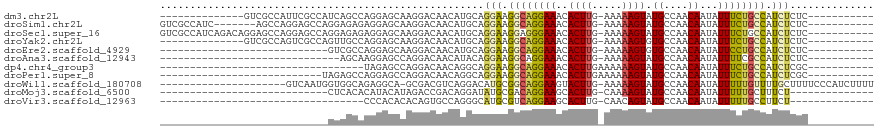
>dm3.chr2L 11941263 93 - 23011544 --------------GUCGCCAUUCGCCAUCAGCCAGGAGCAAGGACAACAUGCAGGAAGGCAGGAAACACUUG-AAAAAGUAUGCCAACAAUAUUUCUGCCAUCUCUC----------- --------------(((((..((.((.....)).))..))...))).....((((((((((((....)((((.-...)))).)))).......)))))))........----------- ( -20.61, z-score = -0.35, R) >droSim1.chr2L 11748297 100 - 22036055 GUCGCCAUC-------AGCCAGGAGCCAGGAGAGAGGAGCAAGGACAACAUGCAGGAAGGCAGGAAACACUUG-AAAAAGUAUGCCAACAAUAUUUCUGCCAUCUCUC----------- (.(.((...-------.....)).).)..((((((((.(((.(.....).))).....(((((....)((((.-...)))).)))).............)).))))))----------- ( -24.80, z-score = -0.43, R) >droSec1.super_16 140468 107 - 1878335 GUCGCCAUCAGACAGGAGCCAGGAGCCAGGAGAGAGGAGCAAGGACAACAUGCAGGAAGGAGGGAAACACUUG-AAAAAGUAUGCCAACAAUAUUUCUGCCAUCUCUC----------- (((.......))).((..(...)..))..((((((((.(((.(.....).)))(((((.(..((....((((.-...))))...))..)....))))).)).))))))----------- ( -24.20, z-score = 0.29, R) >droYak2.chr2L 8362014 93 - 22324452 --------------GUCGCCAGUCGCCAGUUGCCAGGAGCAAGGACAACAUGCAGGAAGGCAGGAAACACUUG-AAAAAGUGUGCCAACAAUAUUUCUGCCAUCUCUC----------- --------------..........((..((((((........)).))))..)).(((.((((((((((((((.-...))))))..........)))))))).)))...----------- ( -26.30, z-score = -1.00, R) >droEre2.scaffold_4929 13171817 79 + 26641161 ----------------------------GUCGCCAGGAGCAAGGACAACAUGCAGGAAGGCAGGAAACACUUG-AAAAAGUGUGCCAACAAUAUUCCUGCCAUCUCUC----------- ----------------------------..........(((.(.....).))).(((.((((((((((((((.-...))))))..........)))))))).)))...----------- ( -24.80, z-score = -2.40, R) >droAna3.scaffold_12943 2732931 77 + 5039921 ------------------------------AGCAAGGAGCCAGGACAACAUACAGGAAGGCAGGAAACACUUG-AAAAAGUAUGCCAACAAUAUUUUCGCCAUCUCUC----------- ------------------------------.....((((...((.....(((...(..(((((....)((((.-...)))).))))..)..))).....))..)))).----------- ( -14.90, z-score = -1.06, R) >dp4.chr4_group3 9265843 74 - 11692001 ----------------------------------UAGAGCCAGGACAACAGGCAGGAAGGCAGGAAACACUUGAAAAAAGUAUGCCAACAAUAUUUCUGCCAUCUCGC----------- ----------------------------------..(((...(.....).(((((((((((((....)((((.....)))).)))).......))))))))..)))..----------- ( -20.41, z-score = -2.75, R) >droPer1.super_8 458163 81 - 3966273 ---------------------------UAGAGCCAGGAGCCAGGACAACAGGCAGGAAGGCAGGAAACACUUGAAAAAAGUAUGCCAACAAUAUUUCUGCCAUCUCGC----------- ---------------------------..(((((.(....).))......(((((((((((((....)((((.....)))).)))).......))))))))..)))..----------- ( -22.51, z-score = -2.33, R) >droWil1.scaffold_180708 2635878 96 - 12563649 ---------------------GUCAAUGGUGGCAGAGGCA-GCGACGUCAGGACAUGCGGCAGGAAGUACUUG-AAAAAGUAUGCCAACAAUAUUUUUGUUUUGCUUUUCCCAUCUUUU ---------------------......(((((.(((((((-(((..((....)).)))((((.....(((((.-...)))))))))(((((.....))))).))))))).))))).... ( -26.60, z-score = -1.20, R) >droMoj3.scaffold_6500 15558624 76 + 32352404 ----------------------------CUCACACAUACAUAGACCGACAGGAUAUGCGACAGGAAGCACUUG-CAAAAGUAUGCCAACAAUAUUUUUGCUUUCU-------------- ----------------------------................((....))...(((........)))...(-(((((((((.......)))))))))).....-------------- ( -13.20, z-score = -1.22, R) >droVir3.scaffold_12963 6957437 70 + 20206255 ----------------------------------CCCACACACAGUGCCAGGGCAUGCGUCAGGAAGCACUUG-CAACAGUAUGCCAACAAUAUUUUUGCCUUCU-------------- ----------------------------------..(((.....)))....(((((((((((((.....))))-..)).)))))))...................-------------- ( -13.80, z-score = 0.69, R) >consensus ____________________________GUAGCCAGGAGCAAGGACAACAUGCAGGAAGGCAGGAAACACUUG_AAAAAGUAUGCCAACAAUAUUUCUGCCAUCUCUC___________ ......................................................(((.((((((((..((((.....)))).((....))...)))))))).))).............. ( -9.33 = -8.99 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:56 2011