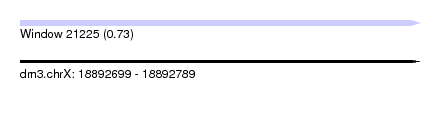
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,892,699 – 18,892,789 |
| Length | 90 |
| Max. P | 0.734178 |

| Location | 18,892,699 – 18,892,789 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 65.29 |
| Shannon entropy | 0.68105 |
| G+C content | 0.34757 |
| Mean single sequence MFE | -15.17 |
| Consensus MFE | -5.87 |
| Energy contribution | -5.82 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
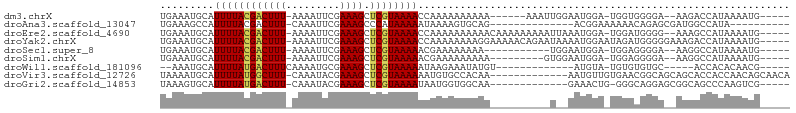
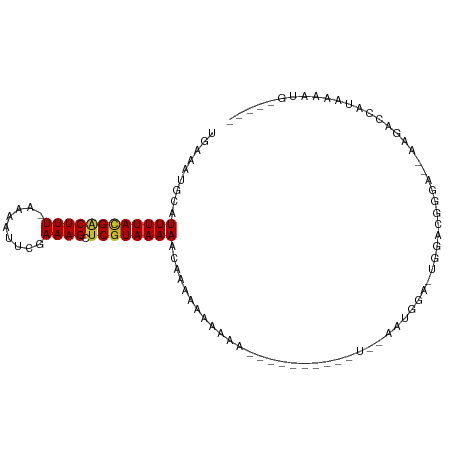
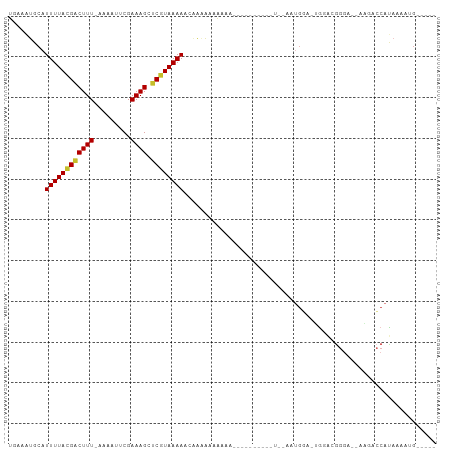
>dm3.chrX 18892699 90 + 22422827 UGAAAUGCAUUUUACGACUUU-AAAAUUCGAAAGCUCGUAAAACCAAAAAAAAAA------AAAUUGGAAUGGA-UGGUGGGGA--AAGACCAUAAAAUG----- ....((.((((((((((((((-........)))).))))))..((((........------...)))))))).)-).((((...--....))))......----- ( -14.60, z-score = -1.65, R) >droAna3.scaffold_13047 1774395 80 + 1816235 UGAAAGCCAUUUUACGACUUU-CAAAUUCGAAAGCCCAUAAAAAUAAAAGUGCAG--------------ACGGAAAAAACAGAGCGAUGGCCAUA---------- .....((((((......((((-(......))))).................((..--------------..............))))))))....---------- ( -10.39, z-score = -0.17, R) >droEre2.scaffold_4690 9174225 96 + 18748788 UGAAAUGCAUUUUACGACUUU-AAAAUUCGAAAGCUCGUAAAACCAAAAAAAAAACAAAAAAAAAUUAAAUGGA-UGGAUGGGG--AAAGCCAUAAAAUG----- .........((((((((((((-........)))).))))))))...............................-...((((..--....))))......----- ( -11.70, z-score = -0.83, R) >droYak2.chrX 17498685 99 + 21770863 UGAAAUGCAUUUUACGACUUU-AAAAUUCGAAAGCUCGUAAAACCAAAAAAAAGGAAAAACAGAAUAAAAUGGAAUAGAUGGGGGAAAGACCAUAAAAUG----- .........((((((((((((-........)))).))))))))((........)).......................((((........))))......----- ( -13.30, z-score = -1.47, R) >droSec1.super_8 1187224 85 + 3762037 UGAAAUGCAUUUUACGACUUU-AAAAUUCGAAAGCUCGUAAAAACGAAAAAAAA-----------UGGAAUGGA-UGGAGGGGA--AAGGCCAUAAAAUG----- .........((((((((((((-........)))).))))))))..........(-----------(((..(...-.........--.)..))))......----- ( -11.02, z-score = -0.55, R) >droSim1.chrX 14598469 87 + 17042790 UGAAAUGCAUUUUACGACUUU-AAAAUUCGAAAGCUCGUAAAAACGAAAAAAAAA---------GUGGAAUGGA-UGGAGGGGA--AAGGCCAUAAAAUG----- ....((.((((((((......-......(....).((((....))))........---------)))))))).)-)....((..--....))........----- ( -12.40, z-score = -0.83, R) >droWil1.scaffold_181096 8113712 79 + 12416693 --AAAUGCAUUUUAUGACUUUCAAAAUGCGAAAGCUCGUAAAAAUAAGAAAUAUGU-------------AUGUA-UGUGUGUGC-----ACCACACAACG----- --..((((((((((((((((((.......))))).))))))))...(....).)))-------------))((.-((((((...-----..)))))))).----- ( -18.20, z-score = -2.28, R) >droVir3.scaffold_12726 830600 91 + 2840439 UAAAAUGCAUUUUAUGGCUUU-CAAAUACGAAAGCUCGUAAAAAAUGUGCCACAA-------------AAUGUUGUGAACGGCAGCAGCACCACCAACAGCAACA .....(((.((((((((((((-(......)))))).)))))))...((((.....-------------..(((((....)))))...))))........)))... ( -24.80, z-score = -2.69, R) >droGri2.scaffold_14853 5261782 85 + 10151454 UAAAGUGCAUUUUAUGACUUU-CAAAUACGAAAGCUCGUAAAAUAAUGGUGGCAA-------------GAAACUG-GGGCAGGAGCGGCAGCCCAAGUCG----- .....((((((((((((((((-(......))))).))))))))).......))).-------------...(((.-((((..........)))).)))..----- ( -20.11, z-score = -0.73, R) >consensus UGAAAUGCAUUUUACGACUUU_AAAAUUCGAAAGCUCGUAAAAACAAAAAAAAAA__________U__AAUGGA_UGGACGGGA__AAGACCAUAAAAUG_____ .........((((((((((((.........)))).)))))))).............................................................. ( -5.87 = -5.82 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:58:41 2011