
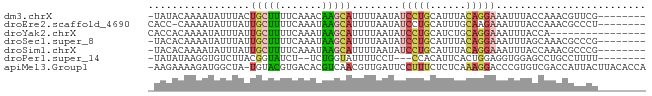
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,880,014 – 18,880,088 |
| Length | 74 |
| Max. P | 0.986212 |

| Location | 18,880,014 – 18,880,088 |
|---|---|
| Length | 74 |
| Sequences | 7 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 61.25 |
| Shannon entropy | 0.74844 |
| G+C content | 0.33813 |
| Mean single sequence MFE | -12.27 |
| Consensus MFE | -4.21 |
| Energy contribution | -4.44 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.70 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943070 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
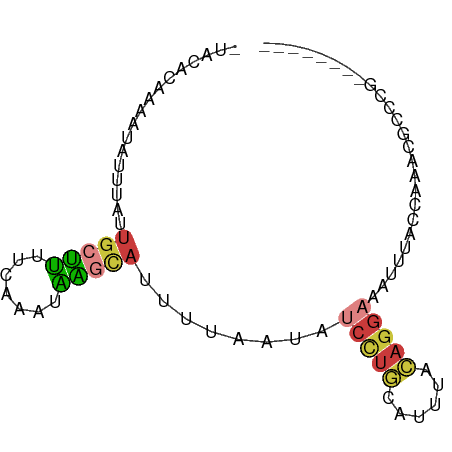
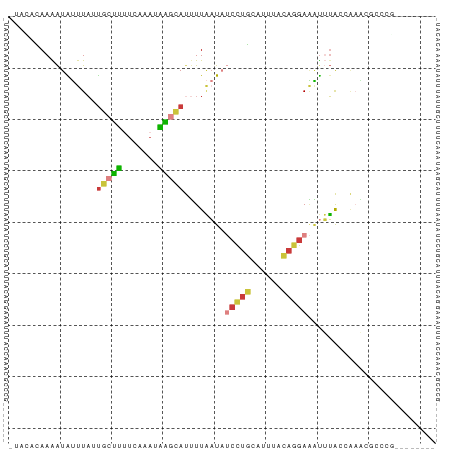
>dm3.chrX 18880014 74 + 22422827 -UAUACAAAAUAUUUACUGCUUUUCAAACAAGCAUUUUAAUAUCCUGCAUUUACAGGAAAUUUACCAAACGUUCG-------- -.........((((...(((((.......)))))....))))(((((......))))).................-------- ( -9.20, z-score = -2.36, R) >droEre2.scaffold_4690 9162079 74 + 18748788 CACC-CAAAAUAUUUAUUGCUUUUCAAAUAAGCAUUUUAAUAUCCUGCAUUUGCAAGAAAUUUACCAAACGCCCU-------- ....-.....((((...(((((.......)))))....))))((.(((....))).)).................-------- ( -8.00, z-score = -1.34, R) >droYak2.chrX 17488226 67 + 21770863 CACCACAAAAUAUUUAUUGCUUUUCAAAUAAGCAUUUUAAUAUCCUGCAUCUGCAGGAAAUUUACCA---------------- ..........((((...(((((.......)))))....))))((((((....)))))).........---------------- ( -14.50, z-score = -3.55, R) >droSec1.super_8 1174740 74 + 3762037 -UACACAAAAUAUUUAUUGCUUUUCAAAUAAGCAUUUUAAUAUCCUGCAUUUACAGGAAAUUUAGCAAACGCCCG-------- -.........((((...(((((.......)))))....))))(((((......)))))......((....))...-------- ( -10.90, z-score = -1.68, R) >droSim1.chrX 14591002 74 + 17042790 -UACACAAAAUAUUUAUUGCUUUUCAAAUAAGCAUUUUAAUAUCCUGCAUUUACAGGAAAUUUACCAAACGCCCG-------- -.........((((...(((((.......)))))....))))(((((......))))).................-------- ( -10.60, z-score = -2.60, R) >droPer1.super_14 1184 69 + 2168203 -UAUAUAAGGUGUCUUACGGUAUCU--UCUGGUAUUUUCCU---CCACAUUCACUGGAGGUGGAGCCUGCCUUUU-------- -.....(((((.......((((((.--...))))))...((---((((.(((....)))))))))...)))))..-------- ( -16.70, z-score = -0.67, R) >apiMel3.Group1 24945645 81 + 25854376 -AAGAAAAGAUGGCUA-UGUACGUGACACGUCAACGUUGAUUCCUUUCUCUCAAAGGACCCGUGUCGACCAUUACUUACACCA -.......(((((...-......((((((((((....))).((((((.....))))))..))))))).))))).......... ( -16.00, z-score = -0.23, R) >consensus _UACACAAAAUAUUUAUUGCUUUUCAAAUAAGCAUUUUAAUAUCCUGCAUUUACAGGAAAUUUACCAAACGCCCG________ .................(((((.......)))))........(((((......)))))......................... ( -4.21 = -4.44 + 0.23)
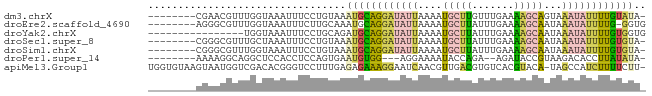
| Location | 18,880,014 – 18,880,088 |
|---|---|
| Length | 74 |
| Sequences | 7 |
| Columns | 83 |
| Reading direction | reverse |
| Mean pairwise identity | 61.25 |
| Shannon entropy | 0.74844 |
| G+C content | 0.33813 |
| Mean single sequence MFE | -15.96 |
| Consensus MFE | -7.34 |
| Energy contribution | -7.80 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.76 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.986212 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 18880014 74 - 22422827 --------CGAACGUUUGGUAAAUUUCCUGUAAAUGCAGGAUAUUAAAAUGCUUGUUUGAAAAGCAGUAAAUAUUUUGUAUA- --------.........((.......)).....((((((((((((....(((((.......)))))...)))))))))))).- ( -15.10, z-score = -2.06, R) >droEre2.scaffold_4690 9162079 74 - 18748788 --------AGGGCGUUUGGUAAAUUUCUUGCAAAUGCAGGAUAUUAAAAUGCUUAUUUGAAAAGCAAUAAAUAUUUUG-GGUG --------.((((((((....(((.((((((....)))))).))).))))))))(((..(((...........)))..-))). ( -15.50, z-score = -1.74, R) >droYak2.chrX 17488226 67 - 21770863 ----------------UGGUAAAUUUCCUGCAGAUGCAGGAUAUUAAAAUGCUUAUUUGAAAAGCAAUAAAUAUUUUGUGGUG ----------------.....(((.((((((....)))))).)))....(((((.......)))))................. ( -14.20, z-score = -1.59, R) >droSec1.super_8 1174740 74 - 3762037 --------CGGGCGUUUGCUAAAUUUCCUGUAAAUGCAGGAUAUUAAAAUGCUUAUUUGAAAAGCAAUAAAUAUUUUGUGUA- --------.((((((((....(((.((((((....)))))).))).)))))))).........((((........))))...- ( -16.80, z-score = -1.81, R) >droSim1.chrX 14591002 74 - 17042790 --------CGGGCGUUUGGUAAAUUUCCUGUAAAUGCAGGAUAUUAAAAUGCUUAUUUGAAAAGCAAUAAAUAUUUUGUGUA- --------.((((((((....(((.((((((....)))))).))).)))))))).........((((........))))...- ( -16.80, z-score = -2.19, R) >droPer1.super_14 1184 69 - 2168203 --------AAAAGGCAGGCUCCACCUCCAGUGAAUGUGG---AGGAAAAUACCAGA--AGAUACCGUAAGACACCUUAUAUA- --------..((((..((.....((((((.......)))---)))......))...--......(....)...)))).....- ( -12.70, z-score = -0.36, R) >apiMel3.Group1 24945645 81 - 25854376 UGGUGUAAGUAAUGGUCGACACGGGUCCUUUGAGAGAAAGGAAUCAACGUUGACGUGUCACGUACA-UAGCCAUCUUUUCUU- ((((....(((...((.((((((..((((((.....)))))).(((....))))))))))).))).-..)))).........- ( -20.60, z-score = -0.59, R) >consensus ________CGAACGUUUGGUAAAUUUCCUGUAAAUGCAGGAUAUUAAAAUGCUUAUUUGAAAAGCAAUAAAUAUUUUGUGUA_ .................................((((((((((((....(((((.......)))))...)))))))))))).. ( -7.34 = -7.80 + 0.46)
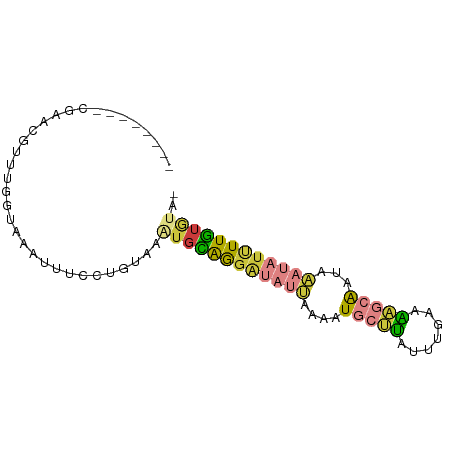
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:58:38 2011