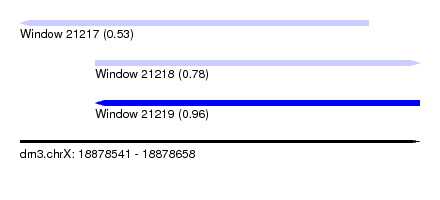
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,878,541 – 18,878,658 |
| Length | 117 |
| Max. P | 0.957555 |

| Location | 18,878,541 – 18,878,643 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 90.75 |
| Shannon entropy | 0.19089 |
| G+C content | 0.38220 |
| Mean single sequence MFE | -20.12 |
| Consensus MFE | -18.04 |
| Energy contribution | -17.95 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.531883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18878541 102 - 22422827 AUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAACACGCAG-CAGGGAACGAAGUCA---------- ...((((...(((((((.....))))(((((((.....)))))))..........(((........)))..............))).-..(....)...))))---------- ( -19.20, z-score = -0.66, R) >droAna3.scaffold_13047 1763041 102 - 1816235 AUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAACACGCAG-CAGGGAGCAAAGUCA---------- ...((((...(((((((.....))))(((((((.....))))))).(((....(((..........................)))..-..))).)))..))))---------- ( -20.77, z-score = -1.11, R) >droEre2.scaffold_4690 9160693 102 - 18748788 AUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAACACGCAG-CAGGGAACGAAGUCA---------- ...((((...(((((((.....))))(((((((.....)))))))..........(((........)))..............))).-..(....)...))))---------- ( -19.20, z-score = -0.66, R) >droYak2.chrX 17486832 102 - 21770863 AUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAACACGCAG-CAGGGAACGAAGUCA---------- ...((((...(((((((.....))))(((((((.....)))))))..........(((........)))..............))).-..(....)...))))---------- ( -19.20, z-score = -0.66, R) >droSec1.super_8 1173292 102 - 3762037 AUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAACACGCAG-CAGGGAACGAAGUCA---------- ...((((...(((((((.....))))(((((((.....)))))))..........(((........)))..............))).-..(....)...))))---------- ( -19.20, z-score = -0.66, R) >droSim1.chrX 14589546 102 - 17042790 AUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAACACGCAG-CAGGGAACGAAGUCA---------- ...((((...(((((((.....))))(((((((.....)))))))..........(((........)))..............))).-..(....)...))))---------- ( -19.20, z-score = -0.66, R) >droWil1.scaffold_181096 8099753 109 - 12416693 AUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGUGACAUGCACAAGUCAAAUAAAUAAACACGCAG-AGGCGUAUAAACGAAAGCAGAG--- ...((((...(((((((.....))))(((((((.....)))))))...............)))...))))...........((((..-..)))).....(....).....--- ( -21.60, z-score = -1.62, R) >droVir3.scaffold_12726 2029108 107 + 2840439 AUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAACACGCAGCCAACCAACCAAACGACUCA------ .((((.(((.(((((((.....))))(((((((.....)))))))........)))..)))))))((((................................))))..------ ( -18.25, z-score = -1.51, R) >droMoj3.scaffold_6473 9406442 103 - 16943266 AUGUGACAUGUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAACACGCAGCCUACCAAACGAGUCA---------- .((((.(((((((((((.....))))(((((((.....)))))))........)).)))))))))......................................---------- ( -22.80, z-score = -2.08, R) >droGri2.scaffold_14853 4906322 113 + 10151454 AUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAACACGCAGCCAGCCAAACGAAUGAGGCAAACUCA .((((......((((((.....))))(((((((.....)))))))........))(((........))).............))))....(((..........)))....... ( -21.80, z-score = -1.04, R) >consensus AUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAACACGCAG_CAGGGAACGAAGUCA__________ .((((......((((((.....))))(((((((.....)))))))........))(((........))).............))))........................... (-18.04 = -17.95 + -0.09)

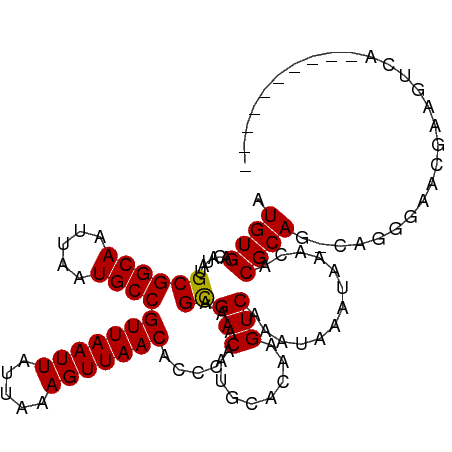
| Location | 18,878,563 – 18,878,658 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 86.70 |
| Shannon entropy | 0.26379 |
| G+C content | 0.36288 |
| Mean single sequence MFE | -21.41 |
| Consensus MFE | -20.01 |
| Energy contribution | -19.83 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.779825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18878563 95 + 22422827 UUUAUUUAUUUGACUUGUGCAUGUCGCUUUUGGGUGUUAACUUUAAUAAUUAACGGCAUUAAUUGCCGCAUAUGUCACAUGUUUGCUUUGCUUCC-------------------------- ..((..(((.((((.(((((....((((....))))..................((((.....))))))))).)))).)))..))..........-------------------------- ( -19.90, z-score = -1.23, R) >droAna3.scaffold_13047 1763063 95 + 1816235 UUUAUUUAUUUGACUUGUGCAUGUCGCUUUUGGGUGUUAACUUUAAUAAUUAACGGCAUUAAUUGCCGCAUAUGUCACAUGUUUGCUUUACCCCG-------------------------- ..((..(((.((((.(((((....((((....))))..................((((.....))))))))).)))).)))..))..........-------------------------- ( -19.90, z-score = -1.32, R) >droEre2.scaffold_4690 9160715 95 + 18748788 UUUAUUUAUUUGACUUGUGCAUGUCGCUUUUGGGUGUUAACUUUAAUAAUUAACGGCAUUAAUUGCCGCAUAUGUCACAUGUUUGCUUUGCGUCC-------------------------- ..((..(((.((((.(((((....((((....))))..................((((.....))))))))).)))).)))..))..........-------------------------- ( -19.90, z-score = -0.78, R) >droYak2.chrX 17486854 95 + 21770863 UUUAUUUAUUUGACUUGUGCAUGUCGCUUUUGGGUGUUAACUUUAAUAAUUAACGGCAUUAAUUGCCGCAUAUGUCACAUGUUUGCUUU--GCCUUU------------------------ ..((..(((.((((.(((((....((((....))))..................((((.....))))))))).)))).)))..))....--......------------------------ ( -19.90, z-score = -1.13, R) >droSec1.super_8 1173314 95 + 3762037 UUUAUUUAUUUGACUUGUGCAUGUCGCUUUUGGGUGUUAACUUUAAUAAUUAACGGCAUUAAUUGCCGCAUAUGUCACAUGUUUGCUUUGCGUCC-------------------------- ..((..(((.((((.(((((....((((....))))..................((((.....))))))))).)))).)))..))..........-------------------------- ( -19.90, z-score = -0.78, R) >droSim1.chrX 14589568 95 + 17042790 UUUAUUUAUUUGACUUGUGCAUGUCGCUUUUGGGUGUUAACUUUAAUAAUUAACGGCAUUAAUUGCCGCAUAUGUCACAUGUUUGCUUUGCGUCC-------------------------- ..((..(((.((((.(((((....((((....))))..................((((.....))))))))).)))).)))..))..........-------------------------- ( -19.90, z-score = -0.78, R) >droWil1.scaffold_181096 8099782 97 + 12416693 UUUAUUUAUUUGACUUGUGCAUGUCACUUUUGGGUGUUAACUUUAAUAAUUAACGGCAUUAAUUGCCGCAUAUGUCACAUGUUCGGCUACAGCGACC------------------------ ......(((.((((.(((((....((((....))))..................((((.....))))))))).)))).)))....((....))....------------------------ ( -21.20, z-score = -1.04, R) >droVir3.scaffold_12726 2029135 120 - 2840439 UUUAUUUAUUUGACUUGUGCAUGUCGCUUUUGGGUGUUAACUUUAAUAAUUAACGGCAUUAAUUGCCGCAUAUGUCACAUGUUUGCUUUC-GCUUUUCCCCGCCAGCUGCAGCCUUCACUU ..........((((.(((((....((((....))))..................((((.....))))))))).))))..(((..(((...-((........)).))).))).......... ( -23.60, z-score = -0.19, R) >droMoj3.scaffold_6473 9406465 108 + 16943266 UUUAUUUAUUUGACUUGUGCAUGUCGCUUUUGGGUGUUAACUUUAAUAAUUAACGGCAUUAAUUGCCGCACAUGUCACAUGUUGGCUUUCGGCUUGUGCCGUUCAAUC------------- .........(..((.(((((((((.((........(((((.........)))))((((.....))))))))))).)))).))..)....((((....)))).......------------- ( -29.50, z-score = -2.37, R) >droGri2.scaffold_14853 4906355 102 - 10151454 UUUAUUUAUUUGACUUGUGCAUGUCGCUUUUGGGUGUUAACUUUAAUAAUUAACGGCAUUAAUUGCCGCAUAUGUCACAUGUUUGCUUUCGGCUUUUCAACC------------------- ......(((.((((.(((((....((((....))))..................((((.....))))))))).)))).)))...((.....)).........------------------- ( -20.40, z-score = -0.95, R) >consensus UUUAUUUAUUUGACUUGUGCAUGUCGCUUUUGGGUGUUAACUUUAAUAAUUAACGGCAUUAAUUGCCGCAUAUGUCACAUGUUUGCUUUCCGCCC__________________________ ......(((.((((.(((((....((((....))))..................((((.....))))))))).)))).)))........................................ (-20.01 = -19.83 + -0.18)
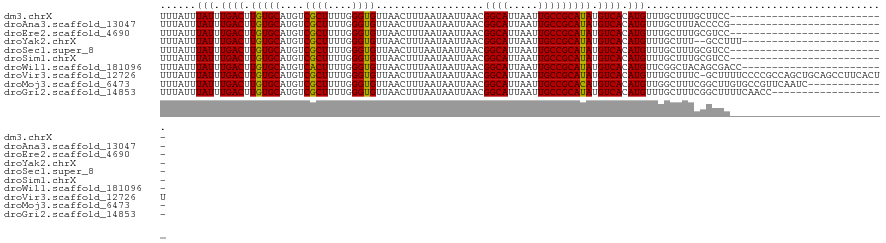
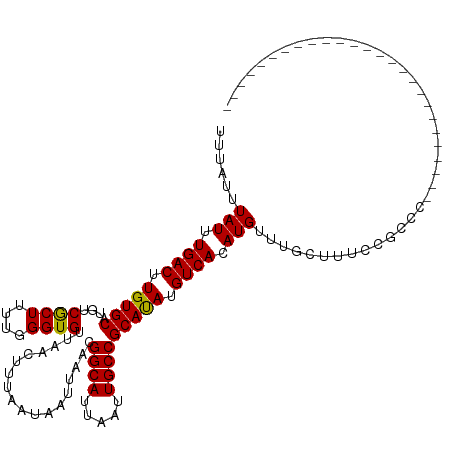
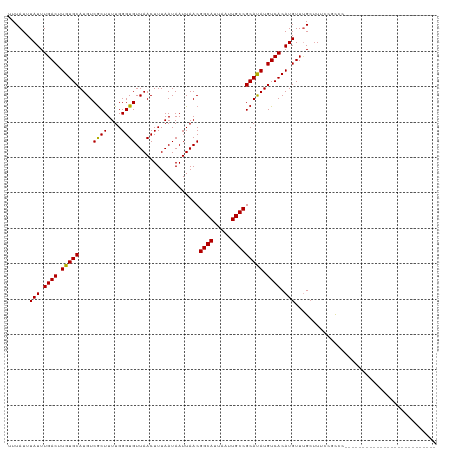
| Location | 18,878,563 – 18,878,658 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 86.70 |
| Shannon entropy | 0.26379 |
| G+C content | 0.36288 |
| Mean single sequence MFE | -22.44 |
| Consensus MFE | -17.95 |
| Energy contribution | -17.86 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18878563 95 - 22422827 --------------------------GGAAGCAAAGCAAACAUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAA --------------------------.............((.((((.(((.(((((((.....))))(((((((.....)))))))........)))..))))))).))............ ( -18.30, z-score = -1.62, R) >droAna3.scaffold_13047 1763063 95 - 1816235 --------------------------CGGGGUAAAGCAAACAUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAA --------------------------..((((...(((.............)))((((.....))))(((((((.....)))))))))))......(((........)))........... ( -19.12, z-score = -1.23, R) >droEre2.scaffold_4690 9160715 95 - 18748788 --------------------------GGACGCAAAGCAAACAUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAA --------------------------.(((((...)).....((((.(((.(((((((.....))))(((((((.....)))))))........)))..))))))).)))........... ( -21.20, z-score = -2.50, R) >droYak2.chrX 17486854 95 - 21770863 ------------------------AAAGGC--AAAGCAAACAUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAA ------------------------...(((--...(....).((((.(((.(((((((.....))))(((((((.....)))))))........)))..))))))).)))........... ( -18.50, z-score = -1.72, R) >droSec1.super_8 1173314 95 - 3762037 --------------------------GGACGCAAAGCAAACAUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAA --------------------------.(((((...)).....((((.(((.(((((((.....))))(((((((.....)))))))........)))..))))))).)))........... ( -21.20, z-score = -2.50, R) >droSim1.chrX 14589568 95 - 17042790 --------------------------GGACGCAAAGCAAACAUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAA --------------------------.(((((...)).....((((.(((.(((((((.....))))(((((((.....)))))))........)))..))))))).)))........... ( -21.20, z-score = -2.50, R) >droWil1.scaffold_181096 8099782 97 - 12416693 ------------------------GGUCGCUGUAGCCGAACAUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGUGACAUGCACAAGUCAAAUAAAUAAA ------------------------.((((((((......))).)))))......((((.....))))(((((((.....))))))).........((((........)))).......... ( -20.70, z-score = -1.23, R) >droVir3.scaffold_12726 2029135 120 + 2840439 AAGUGAAGGCUGCAGCUGGCGGGGAAAAGC-GAAAGCAAACAUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAA ...(((..(.((((..(.((.(((....((-....)).................((((.....))))(((((((.....))))))).)))....)).)..)))))...))).......... ( -29.10, z-score = -1.24, R) >droMoj3.scaffold_6473 9406465 108 - 16943266 -------------GAUUGAACGGCACAAGCCGAAAGCCAACAUGUGACAUGUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAA -------------((((...((((....))))..........((((.(((((((((((.....))))(((((((.....)))))))........)).)))))))))))))........... ( -31.40, z-score = -4.44, R) >droGri2.scaffold_14853 4906355 102 + 10151454 -------------------GGUUGAAAAGCCGAAAGCAAACAUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAA -------------------((((....))))........((.((((.(((.(((((((.....))))(((((((.....)))))))........)))..))))))).))............ ( -23.70, z-score = -2.89, R) >consensus __________________________AGGCGCAAAGCAAACAUGUGACAUAUGCGGCAAUUAAUGCCGUUAAUUAUUAAAGUUAACACCCAAAAGCGACAUGCACAAGUCAAAUAAAUAAA .......................................((.((((.(((.(((((((.....))))(((((((.....)))))))........)))..))))))).))............ (-17.95 = -17.86 + -0.09)
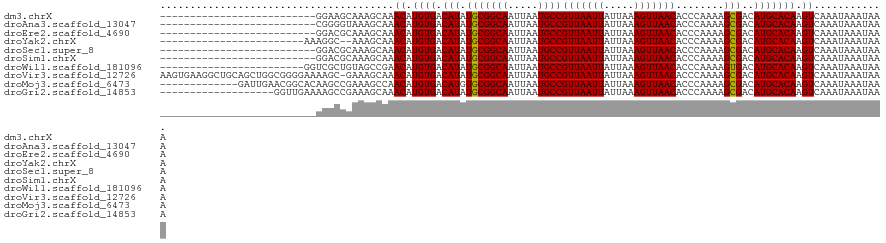
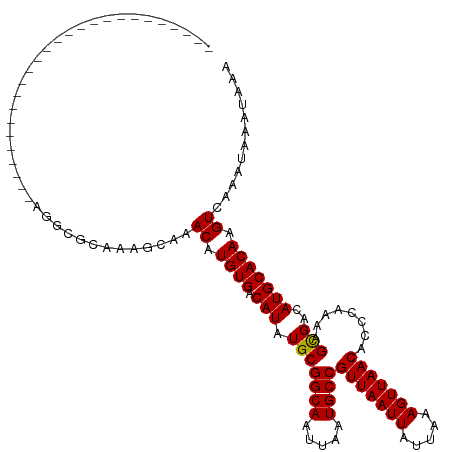
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:58:36 2011