
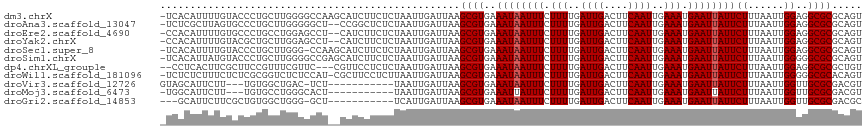
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,875,515 – 18,875,632 |
| Length | 117 |
| Max. P | 0.937660 |

| Location | 18,875,515 – 18,875,631 |
|---|---|
| Length | 116 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.31 |
| Shannon entropy | 0.44056 |
| G+C content | 0.36772 |
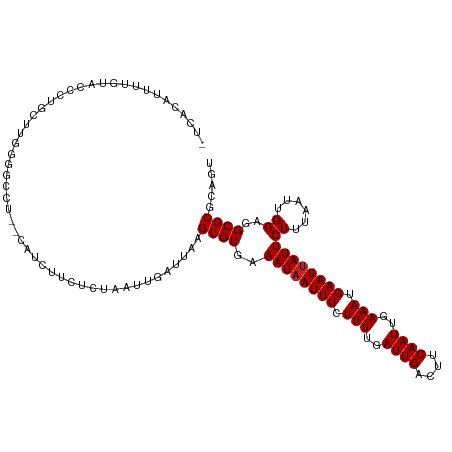
| Mean single sequence MFE | -27.41 |
| Consensus MFE | -15.45 |
| Energy contribution | -15.55 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.935330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18875515 116 + 22422827 -UCACAUUUUGUACCCUGCUUGGGGCCAAGCAUCUUCUCUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGAGGCGCGCAGU -.......((((....((((((....))))))...(((((((((((........(((.((((((.(((..((((....))))..))).))))))))).)))))))))))...)))). ( -29.00, z-score = -1.88, R) >droAna3.scaffold_13047 1760501 114 + 1816235 -UCUCGCUUAGUGCCCUGCUUGGGGGCU--CCGGCUCUCUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGAGGCGCGCAGU -.(((((..(((.(((.....))).)))--...((.((((((((((........(((.((((((.(((..((((....))))..))).))))))))).))))))))))))))).)). ( -34.90, z-score = -2.46, R) >droEre2.scaffold_4690 9157744 114 + 18748788 -CCACAUUUUGUGCCCUGCCUGGAGCCU--CAUCUUCUCUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGAGGCGCGCAGU -.......(((((((((.(((((((...--......)))))...(((((((...(((.((((((.(((..((((....))))..))).))))))))))))))))))))).)))))). ( -28.30, z-score = -2.05, R) >droYak2.chrX 17483869 114 + 21770863 -CCACAUUUUGUACGCUGCUUGGAGCCU--CAUCUUCUCUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGAGGCGCGCAGU -.............(((((((((((...--......))))))........((((..((((((((.(((..((((....))))..))).))))))))((((....))))))))))))) ( -27.00, z-score = -1.69, R) >droSec1.super_8 1170391 115 + 3762037 -UCACAUUUUGUACCCUGCUUGGG-CCAAGCAUCUUCUCUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGAGGCGCGCAGU -.......((((....((((((..-.))))))...(((((((((((........(((.((((((.(((..((((....))))..))).))))))))).)))))))))))...)))). ( -26.00, z-score = -1.21, R) >droSim1.chrX 14586615 116 + 17042790 -UCACAUUAUGUACCCUGCUUGGGGCCGAGCAUCUUCUCUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGGGGCGCGCAGU -.........(..(((((((((....))))))......((((((((........(((.((((((.(((..((((....))))..))).))))))))).)))))))))))..)..... ( -29.30, z-score = -1.57, R) >dp4.chrXL_group1e 10553132 112 + 12523060 --CCUCACUUCGCUUCCGUUUCGUUC---CGUUCCUCUCUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGAGGCGCGCUGU --........((((((((......((---((((..((.......))...)))).))((((((((.(((..((((....))))..))).)))))))).......))))))))...... ( -25.50, z-score = -2.43, R) >droWil1.scaffold_181096 8096526 115 + 12416693 -UCUCUCUUUCUCUCGCGGUCUCUCCAU-CGCUUCCUCUUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGGGGCGCACAGU -..............(((.((((..((.-(((((..((......))..))))))).((((((((.(((..((((....))))..))).))))))))........)))).)))..... ( -29.00, z-score = -3.24, R) >droVir3.scaffold_12726 808524 102 + 2840439 GUAGCAUUCUU---UGUGGCUGAC-UCU-----------UAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGUUGCGCGACGU (((((......---....(((...-((.-----------.....))...)))....((((((((.(((..((((....))))..))).)))))))).........)))))....... ( -20.30, z-score = -1.05, R) >droMoj3.scaffold_6473 7532729 102 - 16943266 -UGGCAUUCUU---UGUGCCUGGGCACU-----------UAAUUGAUUAAGCGUGAAAUUAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGUUGCGCGACGU -.(((((....---.)))))((.(((.(-----------(((((((...((.((((..(((((((..((((......))))..))))))).)))).)))))))))).))).)).... ( -25.80, z-score = -2.65, R) >droGri2.scaffold_14853 5242539 102 + 10151454 ---GCAUUCUUCGCUGUGGCUGGG-GCU-----------UCAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGUUGCGCGACGC ---.......((((.(..((..(.-(((-----------(........))))....((((((((.(((..((((....))))..))).))))))))......)..))..)))))... ( -26.40, z-score = -2.01, R) >consensus _UCACAUUUUGUACCCUGCUUGGGGCCU__CAUCUUCUCUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGAGGCGCGCAGU ..................................................((((..((((((((.(((..((((....))))..))).))))))))((......))..))))..... (-15.45 = -15.55 + 0.09)
| Location | 18,875,515 – 18,875,632 |
|---|---|
| Length | 117 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.32 |
| Shannon entropy | 0.44265 |
| G+C content | 0.37112 |
| Mean single sequence MFE | -27.36 |
| Consensus MFE | -15.45 |
| Energy contribution | -15.55 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.937660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18875515 117 + 22422827 UCACAUUUUGUACCCUGCUUGGGGCCAAGCAUCUUCUCUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGAGGCGCGCAGUC .......((((....((((((....))))))...(((((((((((........(((.((((((.(((..((((....))))..))).))))))))).)))))))))))...)))).. ( -29.00, z-score = -1.86, R) >droAna3.scaffold_13047 1760501 115 + 1816235 UCUCGCUUAGUGCCCUGCUUGGGGGCU--CCGGCUCUCUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGAGGCGCGCAGUC .(((((..(((.(((.....))).)))--...((.((((((((((........(((.((((((.(((..((((....))))..))).))))))))).))))))))))))))).)).. ( -34.90, z-score = -2.57, R) >droEre2.scaffold_4690 9157744 115 + 18748788 CCACAUUUUGUGCCCUGCCUGGAGCCU--CAUCUUCUCUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGAGGCGCGCAGUC .......(((((((((.(((((((...--......)))))...(((((((...(((.((((((.(((..((((....))))..))).))))))))))))))))))))).)))))).. ( -28.30, z-score = -2.08, R) >droYak2.chrX 17483869 115 + 21770863 CCACAUUUUGUACGCUGCUUGGAGCCU--CAUCUUCUCUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGAGGCGCGCAGUC .............(((((((((((...--......))))))........((((..((((((((.(((..((((....))))..))).))))))))((((....))))))))))))). ( -27.10, z-score = -1.74, R) >droSec1.super_8 1170391 116 + 3762037 UCACAUUUUGUACCCUGCUUGGG-CCAAGCAUCUUCUCUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGAGGCGCGCAGUC .......((((....((((((..-.))))))...(((((((((((........(((.((((((.(((..((((....))))..))).))))))))).)))))))))))...)))).. ( -26.00, z-score = -1.19, R) >droSim1.chrX 14586615 117 + 17042790 UCACAUUAUGUACCCUGCUUGGGGCCGAGCAUCUUCUCUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGGGGCGCGCAGUC .........(..(((((((((....))))))......((((((((........(((.((((((.(((..((((....))))..))).))))))))).)))))))))))..)...... ( -29.30, z-score = -1.57, R) >dp4.chrXL_group1e 10553132 113 + 12523060 -CCUCACUUCGCUUCCGUUUCGUUC---CGUUCCUCUCUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGAGGCGCGCUGUC -........((((((((......((---((((..((.......))...)))).))((((((((.(((..((((....))))..))).)))))))).......))))))))....... ( -25.50, z-score = -2.39, R) >droWil1.scaffold_181096 8096526 116 + 12416693 UCUCUCUUUCUCUCGCGGUCUCUCCAU-CGCUUCCUCUUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGGGGCGCACAGUU ..............(((.((((..((.-(((((..((......))..))))))).((((((((.(((..((((....))))..))).))))))))........)))).)))...... ( -29.00, z-score = -3.31, R) >droVir3.scaffold_12726 808525 102 + 2840439 UAGCAUUCUU---UGUGGCUGAC-UCU-----------UAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGUUGCGCGACGUC ..........---.(..(((.((-.((-----------(((....))))).))..((((((((.(((..((((....))))..))).))))))))........)))..)........ ( -19.70, z-score = -0.97, R) >droMoj3.scaffold_6473 7532729 103 - 16943266 UGGCAUUCUU---UGUGCCUGGGCACU-----------UAAUUGAUUAAGCGUGAAAUUAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGUUGCGCGACGUC .(((((....---.)))))((.(((.(-----------(((((((...((.((((..(((((((..((((......))))..))))))).)))).)))))))))).))).))..... ( -25.80, z-score = -2.61, R) >droGri2.scaffold_14853 5242539 103 + 10151454 --GCAUUCUUCGCUGUGGCUGGG-GCU-----------UCAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGUUGCGCGACGCU --.......((((.(..((..(.-(((-----------(........))))....((((((((.(((..((((....))))..))).))))))))......)..))..))))).... ( -26.40, z-score = -1.95, R) >consensus UCACAUUUUGUACCCUGCUUGGGGCCU__CAUCUUCUCUAAUUGAUUAAGCGUGAAAUAAUUUCUUUUGAUUGACUUCAAUUGAAAUGAAUUAUUCUUUAAUUGGAGGCGCGCAGUC .................................................((((..((((((((.(((..((((....))))..))).))))))))((......))..))))...... (-15.45 = -15.55 + 0.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:58:34 2011