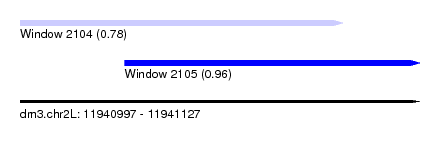
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,940,997 – 11,941,127 |
| Length | 130 |
| Max. P | 0.956153 |

| Location | 11,940,997 – 11,941,102 |
|---|---|
| Length | 105 |
| Sequences | 9 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 70.72 |
| Shannon entropy | 0.55942 |
| G+C content | 0.55487 |
| Mean single sequence MFE | -32.21 |
| Consensus MFE | -15.69 |
| Energy contribution | -17.47 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.777125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
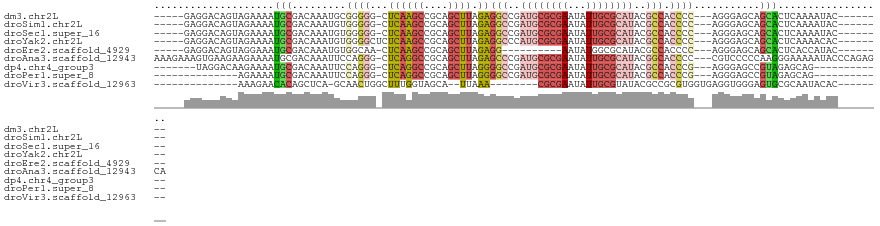
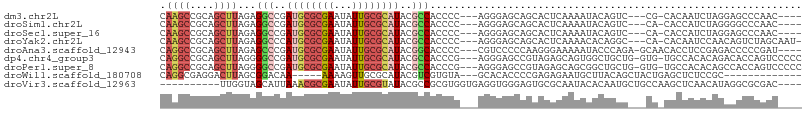
>dm3.chr2L 11940997 105 + 23011544 -----GAGGACAGUAGAAAAUGCGACAAAUGCGGGGG-CUCAAGCCGCAGCUUAGAGGCCGAUGCGCGAAUAUUGCGCAUACGCCACCCC---AGGGAGCAGCACUCAAAAUAC-------- -----(((..(.(((.....)))......(((((((.-(((((((....)))).))).)).((((((((...))))))))...)).((..---..)).))))..))).......-------- ( -34.20, z-score = -1.56, R) >droSim1.chr2L 11748031 105 + 22036055 -----GAGGACAGUAGAAAAUGCGACAAAUGUGGGGG-CUCAAGCCGCAGCUUAGAGGCCGAUGCGCGAAUAUUGCGCAUACGCCACCCC---AGGGAGCAGCACUCAAAAUAC-------- -----(((..(.(((.....)))......(.(((((.-.((((((....)))).))(((..((((((((...))))))))..))).))))---).).....)..))).......-------- ( -33.60, z-score = -1.43, R) >droSec1.super_16 140202 105 + 1878335 -----GAGGACAGUAGAAAAUGCGACAAAUGUGGGGG-CUCAAGCCGCAGCUUAGAGGCCGAUGCGCGAAUAUUGCGCAUACGCCACCCC---AGGGAGCAGCACUCAAAAUAC-------- -----(((..(.(((.....)))......(.(((((.-.((((((....)))).))(((..((((((((...))))))))..))).))))---).).....)..))).......-------- ( -33.60, z-score = -1.43, R) >droYak2.chr2L 8361692 106 + 22324452 -----GAGGACAGUAGAAAAUGCGACAAAUGUGGGGCUCUCAAGCCGCAGCUUAGAGGCCCAUGCGCGAAUAUUGCGCAUACGCCACCCC---AGGGAGCAGCACUCAAAACAC-------- -----(((..(.((.......(((......(((((.((((.((((....)))))))).)))))((((((...))))))...)))..((..---..)).)).)..))).......-------- ( -36.00, z-score = -2.08, R) >droEre2.scaffold_4929 13171556 95 - 26641161 -----GAGGACAGUAGGAAAUGCGACAAAUGUGGCAA-CUCAAGCCGCAGCUUAGAGG----------AAUAUGGCGCAUACGCCACCCC---AGGGAGCAGCACUCACCAUAC-------- -----(((..(.(((.....)))......((((((..-.....))))))((((...((----------....(((((....)))))..))---...)))).)..))).......-------- ( -27.10, z-score = -1.31, R) >droAna3.scaffold_12943 2732575 118 - 5039921 AAAGAAAGUGAAGAAGAAAAUGCGACAAAUUCCAGGG-CUCAGGCCGCAGCUUAGAGCCCGAUGCGCGAAUAUUGCGCAUACGGCACCCC---CGUCCCCCAAGGGAAAAAUACCCAGAGCA ....................((((((........(((-(((((((....)))).)))))).((((((((...))))))))..........---.)))......(((.......)))...))) ( -35.10, z-score = -2.24, R) >dp4.chr4_group3 9265544 99 + 11692001 -------UAGGACAAGAAAAUGCGACAAAUUCCAGGG-CUCAGGCCGCAGCUUAGGGGCCGAUGCGCGAAUAUUGCGCAUACGCCACCCG---AGGGAGCCGUAGAGCAG------------ -------.............(((.((...((((..((-(....)))....((..(((((..((((((((...))))))))..))).))..---))))))..))...))).------------ ( -32.80, z-score = -0.91, R) >droPer1.super_8 457869 92 + 3966273 --------------AGAAAAUGCGACAAAUUCCAGGG-CUCAGGCCGCAGCUUAGGGGCCGAUGCGCGAAUAUUGCGCAUACGCCACCCG---AGGGAGCCGUAGAGCAG------------ --------------......(((.((...((((..((-(....)))....((..(((((..((((((((...))))))))..))).))..---))))))..))...))).------------ ( -32.80, z-score = -1.04, R) >droVir3.scaffold_12963 6957250 89 - 20206255 --------------AAAGAACACAGCUCA-GCAACUGGCUUUGGUAGCA--UUAAA--------CGCGAAUAUUGCGUAUACGCCGCGUGGUGAGGUGGGAGUGCGCAAUACAC-------- --------------......((.(((.((-(...)))))).))...((.--.....--------.))...((((((((((.(.((((.(....).))))).))))))))))...-------- ( -24.70, z-score = 0.04, R) >consensus _____GAGGACAGUAGAAAAUGCGACAAAUGCGGGGG_CUCAAGCCGCAGCUUAGAGGCCGAUGCGCGAAUAUUGCGCAUACGCCACCCC___AGGGAGCAGCACUCAAAAUAC________ .....................((..(......((((...((((((....)))).))(((..((((((((...))))))))..))).)))).....)..))...................... (-15.69 = -17.47 + 1.78)
| Location | 11,941,031 – 11,941,127 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 59.80 |
| Shannon entropy | 0.84272 |
| G+C content | 0.58973 |
| Mean single sequence MFE | -30.77 |
| Consensus MFE | -11.65 |
| Energy contribution | -12.09 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
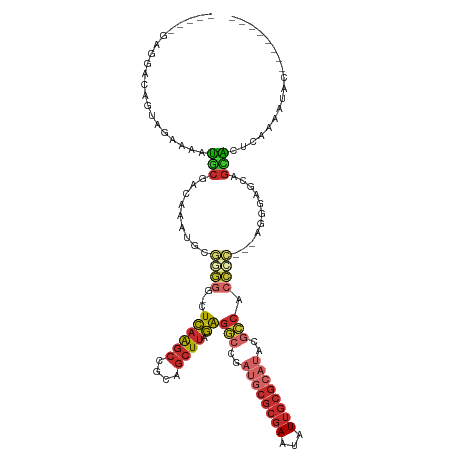
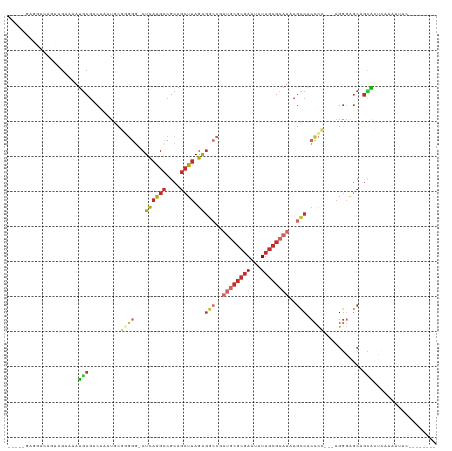
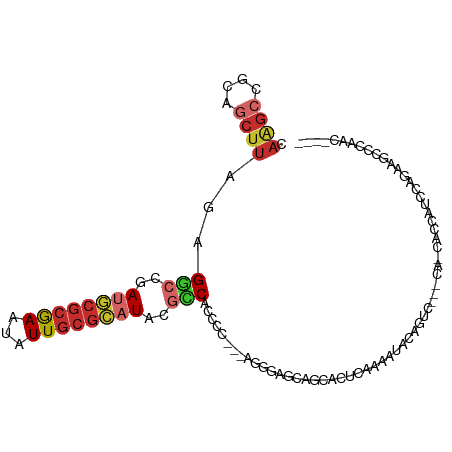
>dm3.chr2L 11941031 96 + 23011544 CAAGCCGCAGCUUAGAGGCCGAUGCGCGAAUAUUGCGCAUACGCCACCCC---AGGGAGCAGCACUCAAAAUACAGUC---CG-CACAAUCUAGGAGCCCAAC---- ......((..(((((((((..((((((((...))))))))..))).((..---..)).((.(.(((........))))---.)-)....)))))).)).....---- ( -26.30, z-score = -1.10, R) >droSim1.chr2L 11748065 96 + 22036055 CAAGCCGCAGCUUAGAGGCCGAUGCGCGAAUAUUGCGCAUACGCCACCCC---AGGGAGCAGCACUCAAAAUACAGUC---CA-CACCAUCUAGGGGCCCAAC---- ...(((((.(((....(((..((((((((...))))))))..))).((..---..))))).))...............---..-..((.....))))).....---- ( -28.50, z-score = -1.45, R) >droSec1.super_16 140236 96 + 1878335 CAAGCCGCAGCUUAGAGGCCGAUGCGCGAAUAUUGCGCAUACGCCACCCC---AGGGAGCAGCACUCAAAAUACAGUC---CA-CACCAUCUAGGAGCCCAAC---- ...((.((.(((....(((..((((((((...))))))))..))).((..---..))))).)).............((---(.-.........))))).....---- ( -24.70, z-score = -1.14, R) >droYak2.chr2L 8361727 99 + 22324452 CAAGCCGCAGCUUAGAGGCCCAUGCGCGAAUAUUGCGCAUACGCCACCCC---AGGGAGCAGCACUCAAAACACAGGC---CA-CACAAUCCAACAGUCUAGCAAU- ...((.((.(((....(((..((((((((...))))))))..))).((..---..))))).))...........((((---..-............)))).))...- ( -25.14, z-score = -1.22, R) >droAna3.scaffold_12943 2732614 99 - 5039921 CAGGCCGCAGCUUAGAGCCCGAUGCGCGAAUAUUGCGCAUACGGCACCCC---CGUCCCCCAAGGGAAAAAUACCCAGA-GCAACACCUCCGAGACCCCCGAU---- .(((..(..((((...(((..((((((((...))))))))..))).....---..((((....))))..........))-))..).)))..............---- ( -25.40, z-score = -1.74, R) >dp4.chr4_group3 9265576 102 + 11692001 CAGGCCGCAGCUUAGGGGCCGAUGCGCGAAUAUUGCGCAUACGCCACCCG---AGGGAGCCGUAGAGCAGUGGCUGCUG-GUG-UGCCACACAGACACCAGUCCCCC ..((((((.((((.(.(((((((((((((...)))))))).))...((..---..)).))).).)))).))))))((((-(((-(.........))))))))..... ( -44.70, z-score = -2.01, R) >droPer1.super_8 457894 102 + 3966273 CAGGCCGCAGCUUAGGGGCCGAUGCGCGAAUAUUGCGCAUACGCCACCCG---AGGGAGCCGUAGAGCAGCGGCUGCUG-GUG-UGCCACACAGCCACCAGUCCCCC ..((((((.((((.(.(((((((((((((...)))))))).))...((..---..)).))).).)))).))))))((((-(((-.((......)))))))))..... ( -48.60, z-score = -2.77, R) >droWil1.scaffold_180708 2635655 86 + 12563649 CAGGCGAGGACUUAGCGGACAA-----AAAAGUUGCGCAUACGUCGUGUA---GCACACCCCGAGAGAAUGCUUACAGCUACUGAGCUCUCCGC------------- ...(((..(((((((..(.(..-----....(((((((.......)))))---))......(....)..........))..))))).))..)))------------- ( -21.20, z-score = 0.53, R) >droVir3.scaffold_12963 6957275 93 - 20206255 ----------UUGGUAGCAUUAAACGCGAAUAUUGCGUAUACGCCGCGUGGUGAGGUGGGAGUGCGCAAUACACAAUGCUGCCAAGCUCAACAUAGGCGCGAC---- ----------(((((((((((......(..((((((((((.(.((((.(....).))))).))))))))))..))))))))))))(((.......))).....---- ( -32.40, z-score = -1.10, R) >consensus CAAGCCGCAGCUUAGAGGCCGAUGCGCGAAUAUUGCGCAUACGCCACCCC___AGGGAGCAGCACUCAAAAUACAGUC___CA_CACCAUCCAGAAGCCCAAC____ .((((....))))...(((..((((((((...))))))))..)))..((......)).................................................. (-11.65 = -12.09 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:33:55 2011