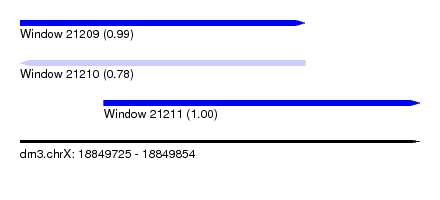
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,849,725 – 18,849,854 |
| Length | 129 |
| Max. P | 0.995797 |

| Location | 18,849,725 – 18,849,817 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 90.23 |
| Shannon entropy | 0.12976 |
| G+C content | 0.47287 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -23.37 |
| Energy contribution | -23.70 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.985733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
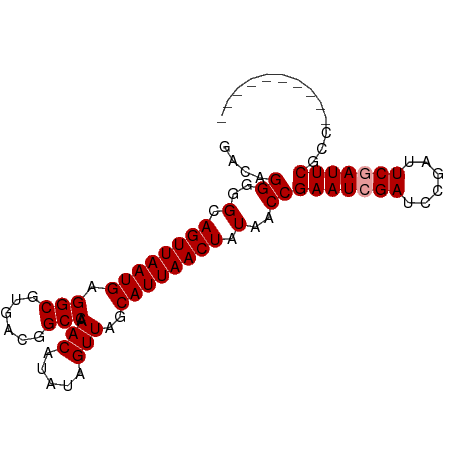
>dm3.chrX 18849725 92 + 22422827 GACAGGGGGCAGUUAAUGAGGCGUGACGGCCAAACAUAUAGUUAGCAUUAACUAUAACCGAAUCGAUCCGAUUCCAUUCGAUUCGUUUCGCC .......(((((((((((.(((......))).(((.....)))..)))))))).....((((((((...........))))))))....))) ( -25.60, z-score = -1.79, R) >droSec1.super_8 1153746 82 + 3762037 GACAGGGGGCAGUUAAUGAGGCGUGACGGCCAAACAUAUAGUUAGCAUUAACUAUAACCGAAUCGAUCCGAUUCGAUUCGCC---------- .......(((((((((((.(((......))).(((.....)))..))))))))......(((((((......))))))))))---------- ( -25.50, z-score = -2.88, R) >droSim1.chrX 14568070 82 + 17042790 GACAGGGGGCAGUUAAUGAGGCGUGACGGCCAAACAUAUAGUUAGCAUUAACUAUAACCGAAUCGAUCCGAUUCGAUUCGCC---------- .......(((((((((((.(((......))).(((.....)))..))))))))......(((((((......))))))))))---------- ( -25.50, z-score = -2.88, R) >consensus GACAGGGGGCAGUUAAUGAGGCGUGACGGCCAAACAUAUAGUUAGCAUUAACUAUAACCGAAUCGAUCCGAUUCGAUUCGCC__________ ....((..(.((((((((.(((......))).(((.....)))..)))))))).)..))(((((((......)))))))............. (-23.37 = -23.70 + 0.33)

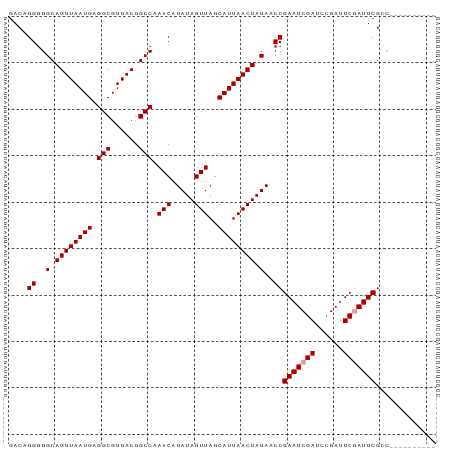
| Location | 18,849,725 – 18,849,817 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 90.23 |
| Shannon entropy | 0.12976 |
| G+C content | 0.47287 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -19.53 |
| Energy contribution | -20.53 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.776688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18849725 92 - 22422827 GGCGAAACGAAUCGAAUGGAAUCGGAUCGAUUCGGUUAUAGUUAAUGCUAACUAUAUGUUUGGCCGUCACGCCUCAUUAACUGCCCCCUGUC (((.(((((((((((((.((......)).))))))))(((((((....))))))).))))).)))........................... ( -24.50, z-score = -1.28, R) >droSec1.super_8 1153746 82 - 3762037 ----------GGCGAAUCGAAUCGGAUCGAUUCGGUUAUAGUUAAUGCUAACUAUAUGUUUGGCCGUCACGCCUCAUUAACUGCCCCCUGUC ----------(((.((((((((((...))))))))))..((((((((..(((.....))).(((......))).)))))))))))....... ( -24.50, z-score = -2.55, R) >droSim1.chrX 14568070 82 - 17042790 ----------GGCGAAUCGAAUCGGAUCGAUUCGGUUAUAGUUAAUGCUAACUAUAUGUUUGGCCGUCACGCCUCAUUAACUGCCCCCUGUC ----------(((.((((((((((...))))))))))..((((((((..(((.....))).(((......))).)))))))))))....... ( -24.50, z-score = -2.55, R) >consensus __________GGCGAAUCGAAUCGGAUCGAUUCGGUUAUAGUUAAUGCUAACUAUAUGUUUGGCCGUCACGCCUCAUUAACUGCCCCCUGUC ..........(((.((((((((((...))))))))))..((((((((..(((.....))).(((......))).)))))))))))....... (-19.53 = -20.53 + 1.00)

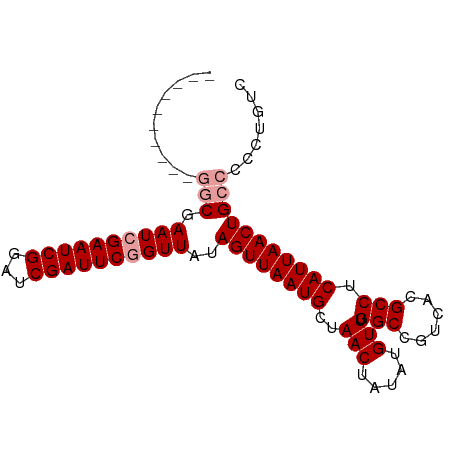
| Location | 18,849,752 – 18,849,854 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 91.22 |
| Shannon entropy | 0.11704 |
| G+C content | 0.48622 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -27.10 |
| Energy contribution | -27.43 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.995797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18849752 102 + 22422827 GGCCAAACAUAUAGUUAGCAUUAACUAUAACCGAAUCGAUCCGAUUCCAUUCGAUUCGUUUCGCCCGCCAGGGAGGCCAGAAUUAAUUGCGGAGCAGCCUGA ((((.....((((((((....))))))))..((((.(((..(((......)))..))).))))(((....))).))))..........((......)).... ( -29.10, z-score = -1.91, R) >droSec1.super_8 1153773 92 + 3762037 GGCCAAACAUAUAGUUAGCAUUAACUAUAACCGAAUCGAUCCGAUUCGAUUCG----------CCCGCCAGGGAGGCCAGAAUUAAUUGCCGAGCAGCCAGA ((((.....((((((((....))))))))..((((((((......))))))))----------(((....))).))))........((((...))))..... ( -29.20, z-score = -3.70, R) >droSim1.chrX 14568097 92 + 17042790 GGCCAAACAUAUAGUUAGCAUUAACUAUAACCGAAUCGAUCCGAUUCGAUUCG----------CCCGCCAGGGAGGCCAGAAUUAAUUGCCGAGCAGCCAGA ((((.....((((((((....))))))))..((((((((......))))))))----------(((....))).))))........((((...))))..... ( -29.20, z-score = -3.70, R) >consensus GGCCAAACAUAUAGUUAGCAUUAACUAUAACCGAAUCGAUCCGAUUCGAUUCG__________CCCGCCAGGGAGGCCAGAAUUAAUUGCCGAGCAGCCAGA ((((.....((((((((....))))))))..((((((((......))))))))..........(((....))).))))........((((...))))..... (-27.10 = -27.43 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:58:29 2011