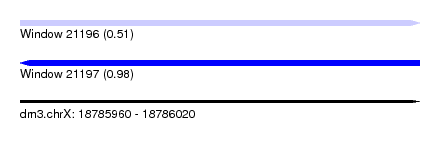
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,785,960 – 18,786,020 |
| Length | 60 |
| Max. P | 0.979638 |

| Location | 18,785,960 – 18,786,020 |
|---|---|
| Length | 60 |
| Sequences | 4 |
| Columns | 64 |
| Reading direction | forward |
| Mean pairwise identity | 74.66 |
| Shannon entropy | 0.40005 |
| G+C content | 0.37326 |
| Mean single sequence MFE | -14.22 |
| Consensus MFE | -5.04 |
| Energy contribution | -6.35 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.507710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
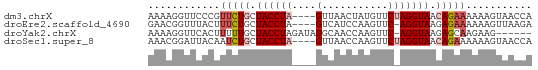
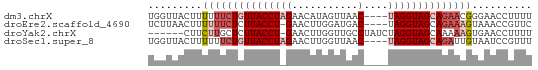
>dm3.chrX 18785960 60 + 22422827 AAAAGGUUCCCGUUCUGCUACCUA----GUUAACUAUGUUCUAGGUAACAGAAAAAAGUAACCA ....(((((...(((((.((((((----(...........))))))).)))))....).)))). ( -18.20, z-score = -4.24, R) >droEre2.scaffold_4690 9078862 59 + 18748788 GAACGGUUUACUUUCUGCUACCUA----GUCAUCCAAGUUC-AGGUAAGAGAAAAAAGUUAAGA ...........(((((..(((((.----..(......)...-)))))..))))).......... ( -7.70, z-score = 0.64, R) >droYak2.chrX 17403286 57 + 21770863 AAAAGGUUCACUUUUUGCUACCUAGAUAGGCAACCAAGUUC-AGGUAAGAGCAAGAAG------ ..........(((((((((((((..((.(....)...))..-))))...)))))))))------ ( -14.60, z-score = -2.22, R) >droSec1.super_8 1088643 60 + 3762037 AAACGGAUUACAAUCUGCUACCUA----GUUAACCAAGUUCUAGGUAACAGAAAAAAGUAACCA ....((.((((..((((.((((((----(...........))))))).)))).....)))))). ( -16.40, z-score = -4.09, R) >consensus AAAAGGUUCACUUUCUGCUACCUA____GUCAACCAAGUUC_AGGUAACAGAAAAAAGUAACCA ............(((((.((((((.................)))))).)))))........... ( -5.04 = -6.35 + 1.31)
| Location | 18,785,960 – 18,786,020 |
|---|---|
| Length | 60 |
| Sequences | 4 |
| Columns | 64 |
| Reading direction | reverse |
| Mean pairwise identity | 74.66 |
| Shannon entropy | 0.40005 |
| G+C content | 0.37326 |
| Mean single sequence MFE | -16.52 |
| Consensus MFE | -8.72 |
| Energy contribution | -10.78 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
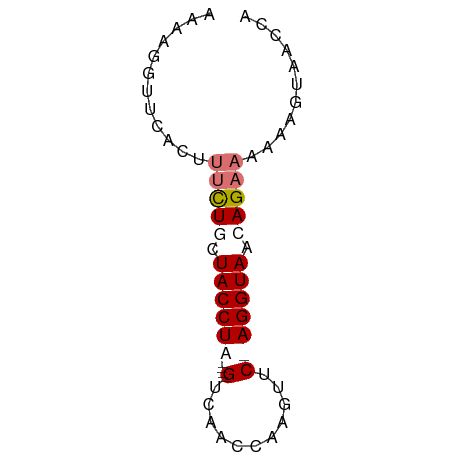
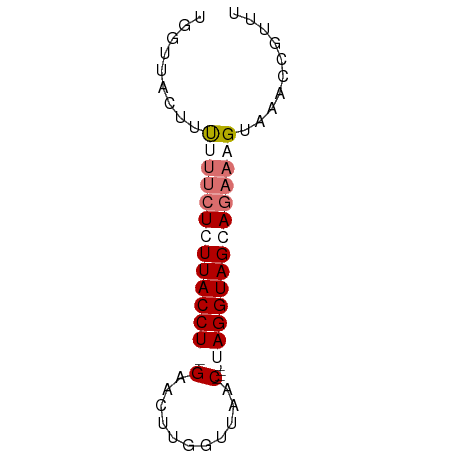
>dm3.chrX 18785960 60 - 22422827 UGGUUACUUUUUUCUGUUACCUAGAACAUAGUUAAC----UAGGUAGCAGAACGGGAACCUUUU .((((.(((..(((((((((((((...........)----)))))))))))).))))))).... ( -22.50, z-score = -4.67, R) >droEre2.scaffold_4690 9078862 59 - 18748788 UCUUAACUUUUUUCUCUUACCU-GAACUUGGAUGAC----UAGGUAGCAGAAAGUAAACCGUUC .........((((((.((((((-(..(......)..----))))))).)))))).......... ( -10.60, z-score = -0.41, R) >droYak2.chrX 17403286 57 - 21770863 ------CUUCUUGCUCUUACCU-GAACUUGGUUGCCUAUCUAGGUAGCAAAAAGUGAACCUUUU ------................-..((((.(((((((....)))))))...))))......... ( -11.40, z-score = -0.99, R) >droSec1.super_8 1088643 60 - 3762037 UGGUUACUUUUUUCUGUUACCUAGAACUUGGUUAAC----UAGGUAGCAGAUUGUAAUCCGUUU .((((((.....((((((((((((...........)----)))))))))))..))))))..... ( -21.60, z-score = -4.74, R) >consensus UGGUUACUUUUUUCUCUUACCU_GAACUUGGUUAAC____UAGGUAGCAGAAAGUAAACCGUUU .........((((((((((((((.................)))))))))))))).......... ( -8.72 = -10.78 + 2.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:58:18 2011