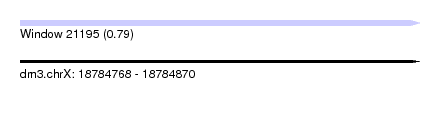
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,784,768 – 18,784,870 |
| Length | 102 |
| Max. P | 0.793051 |

| Location | 18,784,768 – 18,784,870 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.20 |
| Shannon entropy | 0.56755 |
| G+C content | 0.66467 |
| Mean single sequence MFE | -49.13 |
| Consensus MFE | -24.63 |
| Energy contribution | -25.43 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.793051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
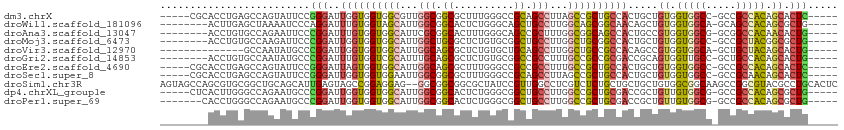
>dm3.chrX 18784768 102 + 22422827 -----CGCACCUGAGCCAGUAUUCCGGGAUUGGUGGUGGCGUUGGCGGCGCUUUGGGCCGCAGCCUUAGCCGCUGCCACUGCUGUGGUGGCC-GCCGCCACAGCACUC----- -----((((((...((((((........))))))))).))).((((((((((..((((....)))).)).)))))))).(((((((((((..-.)))))))))))...----- ( -51.80, z-score = -1.26, R) >droWil1.scaffold_181096 8011664 99 + 12416693 --------ACUUGAGCUAAAAUCCCAGGAUUUGUGGUAGCAUUGGCGGCACUCUGGGCAGCUGCCUUGGCAGCGGCAACAGCUGUGGUGGCA-GCAGCCACAGCGCUG----- --------......(((((((((....)))))....)))).....((((...((((((.((((((.(.((((((....).))))).).))))-)).))).))).))))----- ( -42.00, z-score = -0.40, R) >droAna3.scaffold_13047 1684379 99 + 1816235 --------ACCUGUGCCAGAAUUCCCGGAUUUGUGGUGGCAUUCGCGGCACUUUGGGCAGCCGCUUUGGCGGCAGCCACUGCCGUGGUGGCG-GCGGCCACAACACUG----- --------....(((((.(((((((((......))).)).))))..))))).((((((.((((((.(.(((((((...))))))).).))))-)).))).))).....----- ( -46.50, z-score = -1.35, R) >droMoj3.scaffold_6473 12086912 99 - 16943266 --------ACCUGUGCCAAGAUUCCCGGAUUGGUGGUGGCAUUGGCUGCGCUCUGUGCGGCUGCCUUGGCUGCGGCCACUGCUGUGGUGGCC-GCCGCUACGGCGCUG----- --------......((((.((((....))))..))))(((...((((((((...))))))))(((.((((.(((((((((.....)))))))-)).)))).)))))).----- ( -51.20, z-score = -2.26, R) >droVir3.scaffold_12970 6627075 93 - 11907090 --------------GCCAAUAUGCCCGGAUUGGUGGUGGCAUUGGCAGCGCUCUGUGCUGCAGCCUUGGCUGCCGCCACAGCCGUGGUGGCA-GCUGCUACAGCACUG----- --------------((((..((((((.(.....).).)))))))))........((((((.(((...((((((((((((....)))))))))-)))))).))))))..----- ( -49.40, z-score = -1.92, R) >droGri2.scaffold_14853 553154 99 + 10151454 --------ACCUGUGCCAAUAUGCCCGGAUUUGUGGUCGCAUUUGCAGCGCUCUGUGCGGCCGCCUUUGCCGCCGCGACCGCAGUGGUUGCC-GCUGCCACAGCACUG----- --------....((((...............(((((((((.......((((...))))(((.((....)).))))))))))))(((((....-...))))).))))..----- ( -40.20, z-score = -0.81, R) >droEre2.scaffold_4690 9077664 102 + 18748788 -----CGCACCUGAGCCAGUAUUCCGGGAUUAGUGGUGGCAUUGGCAGCGCUUUGGGCCGCCGCCUUUGCCGCUGCCACUGCUGUGGUGGCC-GCCGCCACAGCACUC----- -----.........((((.((((........)))).))))..(((((((((...((((....))))..).)))))))).(((((((((((..-.)))))))))))...----- ( -48.50, z-score = -1.30, R) >droSec1.super_8 1087463 102 + 3762037 -----CGCACCUGAGCCAGUAUUCCGGGAUUGGUGGUGGAAUUGGCGGCGCUUUGGGCCGCAGCCUUAGCCGCUGCCACUGCUGUGGUGGCC-GCCGCAACAGCACUC----- -----.((.((...((((((........))))))...)).....((((((((..((((....)))).))).((..((((....))))..)).-)))))....))....----- ( -44.20, z-score = -0.05, R) >droSim1.chr3R 4260377 111 - 27517382 AGUAGCCAGCGUGCGGCUGCAGCAUUGAGUAGCCGGAGGAG--GGCGGCGGCGCUAUCCGUUGGCCUCGUCUCUGCUGCUGCUGUGGCGGCAAGCCGGCGUACGCCUGCACUC .((((((.((.(((.((..(((((....(((((.(((.(((--(.((((((......)))))).)))).)).).))))))))))..)).))).)).))).))).......... ( -59.20, z-score = -1.28, R) >dp4.chrXL_group1e 10451455 102 + 12523060 -----CUCACUUGGGCCAGAAUGCCCGGAUUGGUGGUGGCAUUGGCGGCACUCUGGGCGGCUGCCUUGGCCGCUGCGACCGCUGUUGUGGCG-GCCGCCACAGCGCUG----- -----..(((((((((......)))))....))))(((((...((((((.(....)...))))))..((((((..((((....))))..)))-)))))))).......----- ( -52.60, z-score = -1.44, R) >droPer1.super_69 276577 100 - 344178 -------CACCUGGGCCAGAAUGCCCGGAUUGGUGGUGGCAUUGGCGGCACUCUGGGCGGCUGCCUUGGCCGCUGCGACCGCUGUUGUGGCG-GCCGCCACAGCGCUG----- -------...((((((......))))))...(((((((((...((((((.(....)...))))))..((((((..((((....))))..)))-))))))))..)))).----- ( -54.80, z-score = -1.88, R) >consensus ________ACCUGAGCCAGUAUUCCCGGAUUGGUGGUGGCAUUGGCGGCGCUCUGGGCCGCUGCCUUGGCCGCUGCCACUGCUGUGGUGGCC_GCCGCCACAGCACUG_____ ..........................((..((((((((((...(((.((..........)).)))...))))))))))...))((.(((((.....))))).))......... (-24.63 = -25.43 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:58:16 2011