| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,771,291 – 18,771,382 |
| Length | 91 |
| Max. P | 0.982552 |

| Location | 18,771,291 – 18,771,382 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 73.82 |
| Shannon entropy | 0.53906 |
| G+C content | 0.64914 |
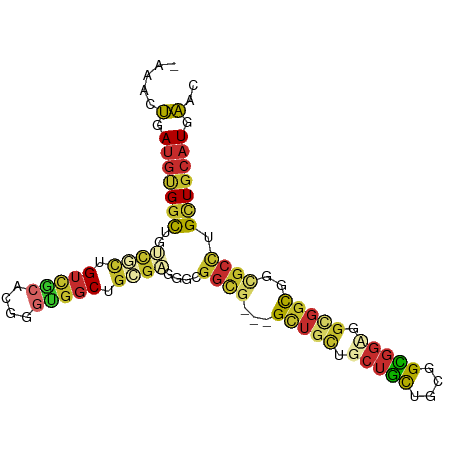
| Mean single sequence MFE | -39.89 |
| Consensus MFE | -26.01 |
| Energy contribution | -26.21 |
| Covariance contribution | 0.20 |
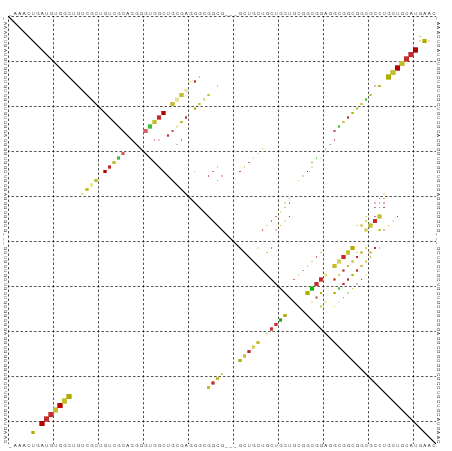
| Combinations/Pair | 1.71 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.982552 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
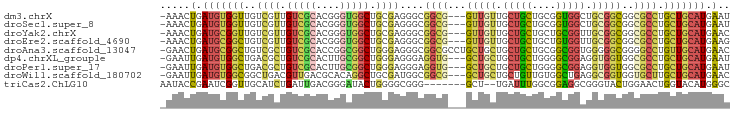
>dm3.chrX 18771291 91 - 22422827 -AAACUGAUGUGGUUGUCGUUGUCGCACGGGUGGCUGCGAGGGCGGCG---GUUGUUGCUGCUGCGGUGGCUGCGGCGGCGCCUGCUGCAUGAAU -....(.(((..((.(.((((((((((...((.((((((..((((((.---...))))))..)))))).)))))))))))).).))..))).).. ( -41.50, z-score = -1.32, R) >droSec1.super_8 1074057 91 - 3762037 -AAACUGAUGUGGUUGUCGUUGUCGCACGGGUGGCUGCGAGGGCGGCG---GUUGUUGCUGCUGCGGUGGCUGCGGCGGCGCCUGCUGCAUGAAU -....(.(((..((.(.((((((((((...((.((((((..((((((.---...))))))..)))))).)))))))))))).).))..))).).. ( -41.50, z-score = -1.32, R) >droYak2.chrX 17388421 91 - 21770863 -AAACUGAUGCGGUUGUCGUUGUCGCACGGGUGGCUGCGAGGGCGGCG---GUUGUUGCUGCUGCUGCGGUUGCGGCGGCGCCUGCUGCAUGAAC -....(.(((((((.(.((((((((((......(((((...(((((((---((....)))))))))))))))))))))))).).))))))).).. ( -44.50, z-score = -1.90, R) >droEre2.scaffold_4690 9064154 91 - 18748788 -AAACUGAUGCGGCUGUCGUUGUCGCACGGGUGGCUGCGAGGGCGGCG---GUUGUUGCUGCUGCUGUGGUUGCGGCGGCGCCUGCUGCAUGAAG -....(.(((((((.((((((.(((((........))))).))))))(---((.((((((((.((....)).))))))))))).))))))).).. ( -44.50, z-score = -1.87, R) >droAna3.scaffold_13047 183329 94 + 1816235 -GAACUGAUGCGGCUGUCGCUGUCGCACCGGCGGCUGGGAGGGCGGCGCCUGCUGCUGCUGCUGCGGCGGUGGGGGCGGGGCCUGUUGCAUGAAC -....(.((((((((.((.(.(((((....))))).).)).)))(((.((((((.(..((((....))))..).)))))))))...))))).).. ( -50.10, z-score = -2.05, R) >dp4.chrXL_group1e 3189886 91 - 12523060 -GAAUUGAUGUGGCUGACGCUGUCGCACUUGCGGCUGGGAGGGAGGUG---GCUGCUGCUGCUGGGGCGGAGGUGGUGGCGCCUGCUGCAUGAAU -..(((.(((..((...(.(.(((((....))))).).).(((..((.---((..((.((((....)))).))..)).)).)))))..))).))) ( -37.70, z-score = -1.65, R) >droPer1.super_17 126987 91 - 1930428 -GAAUUGAUGUGGCUGACGCUGUCGCACUUGCGGCUGGGAGGGAGGUG---GCUGCUGCUGCUGGGGCGGAGGUGGUGGCGCCUGCUGCAUGAAU -..(((.(((..((...(.(.(((((....))))).).).(((..((.---((..((.((((....)))).))..)).)).)))))..))).))) ( -37.70, z-score = -1.65, R) >droWil1.scaffold_180702 4452156 91 - 4511350 -GAAUUGAUGUGGCGGCUGACGUUGACGCACAGGCUGCGAUGGCGGCG---GCUGCUGCUGUUGUGGCUGAGGCGGUGGUGCUUGCUGCAUGAAC -...((.(((..(((((...((....(((...(((..((((((((((.---...))))))))))..)))...))).))..)).)))..))).)). ( -40.60, z-score = -2.81, R) >triCas2.ChLG10 6233889 86 - 8806720 AAUACCGAAUCGGUUGCAUCUGAUUGACGGGAUACUGGGGCGGG-------GCU--UGAUUUGGCGGAGGCGGGUACUGGAACUGGUACAUGGGC ....(((((((((......))))))..)))..(((..(..(.((-------(((--((.(((....))).))))).)).)..)..)))....... ( -20.90, z-score = -0.04, R) >consensus _AAACUGAUGUGGCUGUCGCUGUCGCACGGGUGGCUGCGAGGGCGGCG___GCUGCUGCUGCUGCGGCGGAGGCGGCGGCGCCUGCUGCAUGAAC .....(.(((((((..((((.(((((....))))).))))....((((...(((((.(((((....))))).)))))..)))).))))))).).. (-26.01 = -26.21 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:58:14 2011