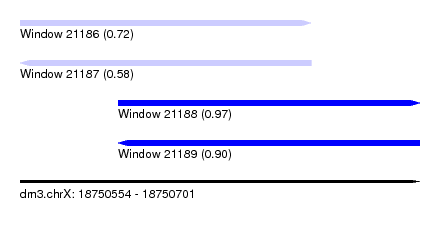
| Sequence ID | dm3.chrX |
|---|---|
| Location | 18,750,554 – 18,750,701 |
| Length | 147 |
| Max. P | 0.969022 |

| Location | 18,750,554 – 18,750,661 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 91.61 |
| Shannon entropy | 0.14345 |
| G+C content | 0.36174 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -23.22 |
| Energy contribution | -22.38 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.724284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18750554 107 + 22422827 AUGCUAAGUUUAAUGCUGAAGGUCAAUUAGAGU-GGGCUACCAGUUGCCCUUUCUUACAAUCUGAUCAUGAUAAUCGUUAUCAUAGUUUAACGAUUC-GCUGAUCGUGA ......(((.....)))..((((.....((((.-((((........)))).))))....))))(((((.(..((((((((........)))))))).-.)))))).... ( -25.90, z-score = -1.25, R) >droEre2.scaffold_4690 9043459 107 + 18748788 AUGCUGAAAUGAAUACUGAAGGUCAAUUAGAGU-GGGUUUUCAGUUGCCCUUUCUUAUAAUCUGAUCAUAAUAAUCGUUAUCAUAGUUUAACGAUUC-GGUGAUCGUGA ........((((.(((((((((((((((((((.-((((........))))...))).)))).))))).......((((((........)))))))))-)))).)))).. ( -28.00, z-score = -2.32, R) >droYak2.chrX 17367522 107 + 21770863 AUGCUAAAAUAAAUACUGAAGGUCAAUUAGAGU-GGGUUUUCAGUUGCCCUUUCUUAUAAUUUGAUCAUGAUAGUCGUUAUCACAGUUUAACGAUUC-GGUGAUCGUGA .............(((((((((((((..((((.-((((........)))).))))......)))))).......((((((........)))))))))-))))....... ( -25.80, z-score = -1.79, R) >droSec1.super_8 1053340 109 + 3762037 AUGCUACAUUUAAUGCUGAAGGUCAAUUAGAGUUGGGCUUCCAGUUGCCUUUUCUUAUAAUCUGAUCAUGAUAAUCGUUAUCAUAGUUUAACGAUUCCGGUGAUCGUGA ....(((.......(((((((.(((((....))))).))).))))..................((((((...((((((((........))))))))...))))))))). ( -25.40, z-score = -1.10, R) >droSim1.chrX_random 4863025 109 + 5698898 AUGCUACAUUUAAUGCUGAAGGUCAAUUAGAGUUGGGCUUCCAGUUGCCUUUUCUUAUAAUCUGAUCAUGAUAAUCGUUAUCAUAGUUUAACGAUUCCGGUGAUCGUGA ....(((.......(((((((.(((((....))))).))).))))..................((((((...((((((((........))))))))...))))))))). ( -25.40, z-score = -1.10, R) >consensus AUGCUAAAUUUAAUGCUGAAGGUCAAUUAGAGU_GGGCUUCCAGUUGCCCUUUCUUAUAAUCUGAUCAUGAUAAUCGUUAUCAUAGUUUAACGAUUC_GGUGAUCGUGA .............(((...((((.....(((...((((........))))..)))....))))((((((...((((((((........))))))))...))))))))). (-23.22 = -22.38 + -0.84)

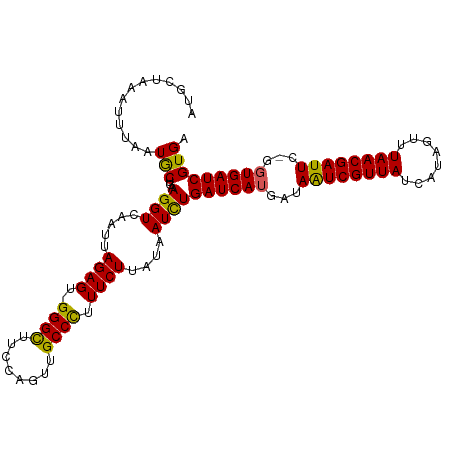
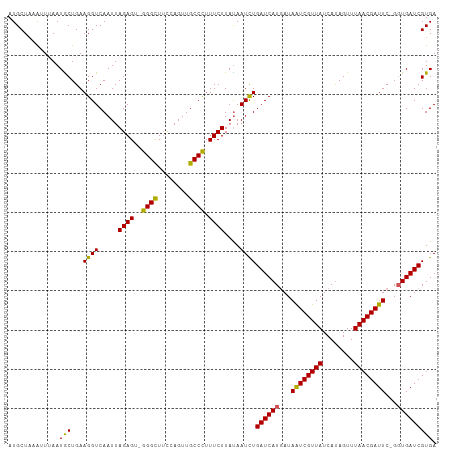
| Location | 18,750,554 – 18,750,661 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 91.61 |
| Shannon entropy | 0.14345 |
| G+C content | 0.36174 |
| Mean single sequence MFE | -20.13 |
| Consensus MFE | -17.50 |
| Energy contribution | -18.06 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18750554 107 - 22422827 UCACGAUCAGC-GAAUCGUUAAACUAUGAUAACGAUUAUCAUGAUCAGAUUGUAAGAAAGGGCAACUGGUAGCCC-ACUCUAAUUGACCUUCAGCAUUAAACUUAGCAU ....(((((..-.((((((((........))))))))....))))).((..((.(((..((((........))))-..))).....))..)).((..........)).. ( -24.00, z-score = -1.99, R) >droEre2.scaffold_4690 9043459 107 - 18748788 UCACGAUCACC-GAAUCGUUAAACUAUGAUAACGAUUAUUAUGAUCAGAUUAUAAGAAAGGGCAACUGAAAACCC-ACUCUAAUUGACCUUCAGUAUUCAUUUCAGCAU ....(((((..-.((((((((........))))))))....))))).........((((.((..((((((.....-.............))))))..)).))))..... ( -19.37, z-score = -1.75, R) >droYak2.chrX 17367522 107 - 21770863 UCACGAUCACC-GAAUCGUUAAACUGUGAUAACGACUAUCAUGAUCAAAUUAUAAGAAAGGGCAACUGAAAACCC-ACUCUAAUUGACCUUCAGUAUUUAUUUUAGCAU ....(((((..-((.((((((........))))))...)).)))))........(((..(((..........)))-..))).((((.....)))).............. ( -15.10, z-score = -0.34, R) >droSec1.super_8 1053340 109 - 3762037 UCACGAUCACCGGAAUCGUUAAACUAUGAUAACGAUUAUCAUGAUCAGAUUAUAAGAAAAGGCAACUGGAAGCCCAACUCUAAUUGACCUUCAGCAUUAAAUGUAGCAU ....(((((....((((((((........))))))))....)))))............(((((((.((((........)))).))).))))..((..........)).. ( -21.10, z-score = -1.56, R) >droSim1.chrX_random 4863025 109 - 5698898 UCACGAUCACCGGAAUCGUUAAACUAUGAUAACGAUUAUCAUGAUCAGAUUAUAAGAAAAGGCAACUGGAAGCCCAACUCUAAUUGACCUUCAGCAUUAAAUGUAGCAU ....(((((....((((((((........))))))))....)))))............(((((((.((((........)))).))).))))..((..........)).. ( -21.10, z-score = -1.56, R) >consensus UCACGAUCACC_GAAUCGUUAAACUAUGAUAACGAUUAUCAUGAUCAGAUUAUAAGAAAGGGCAACUGGAAGCCC_ACUCUAAUUGACCUUCAGCAUUAAAUUUAGCAU ....(((((....((((((((........))))))))....)))))............(((((((.((((........)))).))).)))).................. (-17.50 = -18.06 + 0.56)

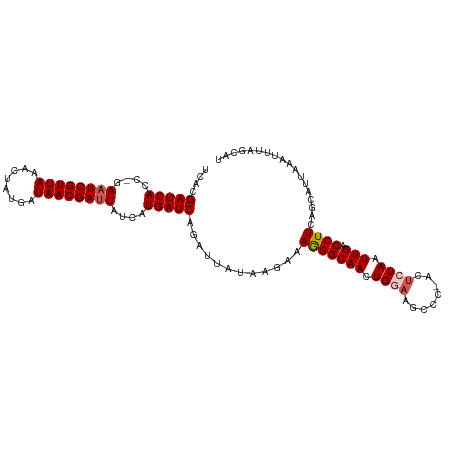
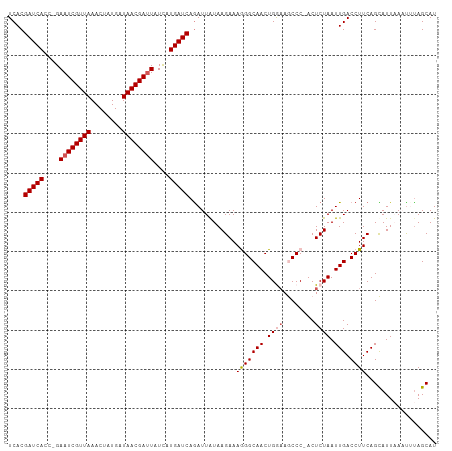
| Location | 18,750,590 – 18,750,701 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 89.26 |
| Shannon entropy | 0.18877 |
| G+C content | 0.33994 |
| Mean single sequence MFE | -24.95 |
| Consensus MFE | -19.50 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18750590 111 + 22422827 CUACCAGUUGCCCUUUCUUACAAUCUGAUCAUGAUAAUCGUUAUCAUAGUUUAACGAUUC-GCUGAUCGUGAUCUUUCAAAAAGCAAGCAGAACUUGUUUUUGAUAAGAACG ..............((((((......(((((((((((((((((........)))))))).-....)))))))))..(((((((.((((.....))))))))))))))))).. ( -28.10, z-score = -3.46, R) >droEre2.scaffold_4690 9043495 110 + 18748788 UUUUCAGUUGCCCUUUCUUAUAAUCUGAUCAUAAUAAUCGUUAUCAUAGUUUAACGAUUC-GGUGAUCGUGAUCUUCCAAAAAGCAA-CAGAACCAACUUUUGAUAAGAACG ..(((.(((((..(((......((((((((((...((((((((........)))))))).-.))))))).))).....)))..))))-).)))................... ( -23.20, z-score = -2.72, R) >droYak2.chrX 17367558 108 + 21770863 UUUUCAGUUGCCCUUUCUUAUAAUUUGAUCAUGAUAGUCGUUAUCACAGUUUAACGAUUC-GGUGAUCGUGAUCUUUCAAAAAACAAG-GGACCUAAUUUUUGAUAAGCA-- .....((...(((((...........(((((((((((((((((........)))))))).-....)))))))))...........)))-))..))...............-- ( -23.65, z-score = -1.78, R) >droSec1.super_8 1053377 112 + 3762037 CUUCCAGUUGCCUUUUCUUAUAAUCUGAUCAUGAUAAUCGUUAUCAUAGUUUAACGAUUCCGGUGAUCGUGAUCUUUUAAAAACCAAACAGAACUUGUUUUUGAUAAGAACG ..............(((((((.....(((((((((.((((..(((..........)))..)))).)))))))))...(((((((............)))))))))))))).. ( -23.90, z-score = -2.44, R) >droSim1.chrX_random 4863062 112 + 5698898 CUUCCAGUUGCCUUUUCUUAUAAUCUGAUCAUGAUAAUCGUUAUCAUAGUUUAACGAUUCCGGUGAUCGUGAUCUUUCAAAAACCAAACAGAACUUGUUUUUGAUAAGAACG ..............((((((......(((((((((.((((..(((..........)))..)))).)))))))))..((((((((............)))))))))))))).. ( -25.90, z-score = -2.99, R) >consensus CUUCCAGUUGCCCUUUCUUAUAAUCUGAUCAUGAUAAUCGUUAUCAUAGUUUAACGAUUC_GGUGAUCGUGAUCUUUCAAAAACCAAACAGAACUUGUUUUUGAUAAGAACG ..............(((((((.....(((((((((((((((((........))))))))......)))))))))...((((((..............))))))))))))).. (-19.50 = -19.98 + 0.48)

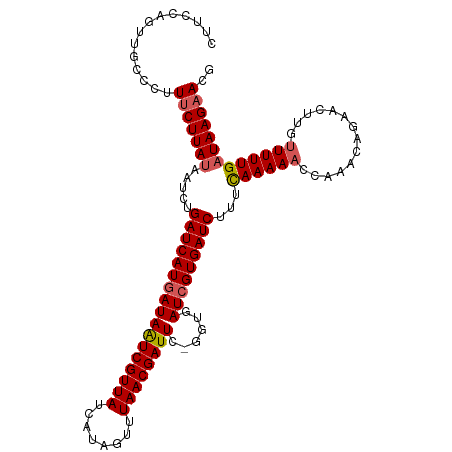
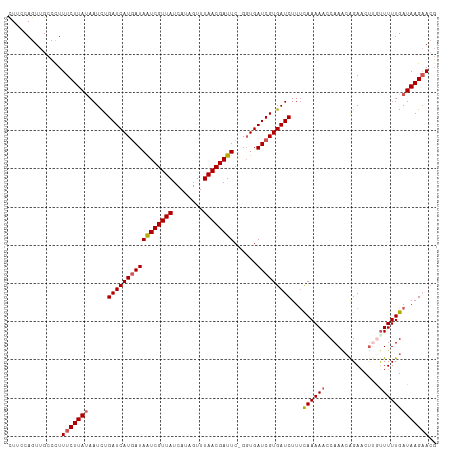
| Location | 18,750,590 – 18,750,701 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 89.26 |
| Shannon entropy | 0.18877 |
| G+C content | 0.33994 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -21.24 |
| Energy contribution | -21.32 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.903757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 18750590 111 - 22422827 CGUUCUUAUCAAAAACAAGUUCUGCUUGCUUUUUGAAAGAUCACGAUCAGC-GAAUCGUUAAACUAUGAUAACGAUUAUCAUGAUCAGAUUGUAAGAAAGGGCAACUGGUAG ..(((((((((((((((((.....)))).)))))))..((((..(((((..-.((((((((........))))))))....))))).)))).)))))).((....))..... ( -29.60, z-score = -2.94, R) >droEre2.scaffold_4690 9043495 110 - 18748788 CGUUCUUAUCAAAAGUUGGUUCUG-UUGCUUUUUGGAAGAUCACGAUCACC-GAAUCGUUAAACUAUGAUAACGAUUAUUAUGAUCAGAUUAUAAGAAAGGGCAACUGAAAA ...................(((.(-(((((((((..(.((((..(((((..-.((((((((........))))))))....))))).)))).)...)))))))))).))).. ( -26.20, z-score = -2.09, R) >droYak2.chrX 17367558 108 - 21770863 --UGCUUAUCAAAAAUUAGGUCC-CUUGUUUUUUGAAAGAUCACGAUCACC-GAAUCGUUAAACUGUGAUAACGACUAUCAUGAUCAAAUUAUAAGAAAGGGCAACUGAAAA --(((((.(((((((..((....-))...)))))))..(((((((((....-..)))))......((((((.....)))))))))).............)))))........ ( -21.40, z-score = -1.51, R) >droSec1.super_8 1053377 112 - 3762037 CGUUCUUAUCAAAAACAAGUUCUGUUUGGUUUUUAAAAGAUCACGAUCACCGGAAUCGUUAAACUAUGAUAACGAUUAUCAUGAUCAGAUUAUAAGAAAAGGCAACUGGAAG ..(((((((...(((((.....)))))...........((((..(((((....((((((((........))))))))....))))).))))))))))).((....))..... ( -22.20, z-score = -1.30, R) >droSim1.chrX_random 4863062 112 - 5698898 CGUUCUUAUCAAAAACAAGUUCUGUUUGGUUUUUGAAAGAUCACGAUCACCGGAAUCGUUAAACUAUGAUAACGAUUAUCAUGAUCAGAUUAUAAGAAAAGGCAACUGGAAG ..((((((((((((((.((......)).))))))))..((((..(((((....((((((((........))))))))....))))).)))).)))))).((....))..... ( -26.30, z-score = -2.23, R) >consensus CGUUCUUAUCAAAAACAAGUUCUGCUUGCUUUUUGAAAGAUCACGAUCACC_GAAUCGUUAAACUAUGAUAACGAUUAUCAUGAUCAGAUUAUAAGAAAGGGCAACUGGAAG ..(((((((((((((((((.....)))).)))))))........(((((....((((((((........))))))))....)))))......)))))).((....))..... (-21.24 = -21.32 + 0.08)

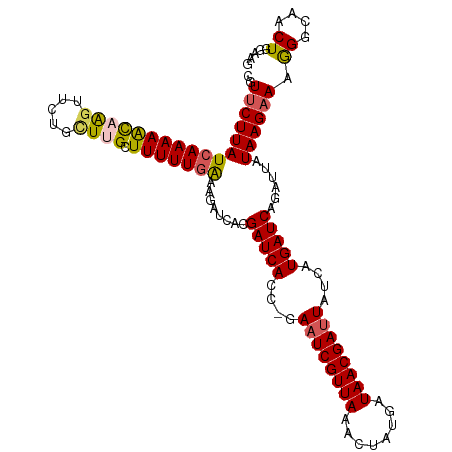
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:58:11 2011